Abstract
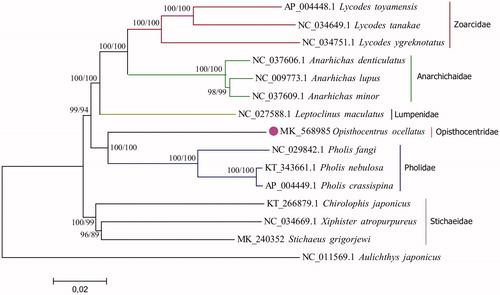
The complete mitochondrial genome (mitogenome) of Opisthocentrus ocellatus (Zoarcales: Opisthocentridae) was sequenced for the first time. The whole genome was 16,525 bp in length and contained 13 protein-coding genes, 22 transfer RNA genes (tRNA), two ribosome (rRNA) genes, and two non-coding regions. The overall base composition of the mitochondrial DNA (mtDNA) was 26.2% for A, 28.3% for T, 27.4% for C, and 18.1% for G. Six protein-coding genes include incomplete stop codons while others have standard ones, and start codons are typically ATG, GTG, ATA, or ATC. The phylogenetic analysis, which based on the mitogenomes of O. ocellatus and other 14 fish species from Zoarcales infraorder indicated that O. ocellatus is more closely related to the family Pholidae than to Stichaeidae.
Keywords:
The ocellated blenny, Opisthocentrus ocellatus, is one of common species of the recently established family Opisthocentridae (Teleostei: Perciformes: Zoarcales) (Radchenko Citation2015; Fricke et al. Citation2018). Although of no commercial value, it is usual in nearshore waters. The ocellated blenny is a typical representative of food chains in boreal communities, dwelling in algae and rocky nearshore habitats, usually at a depth of less than 70 m (Mecklenburg and Sheiko Citation2004). Its range includes the following areas of the North Western Pacific: the western Bering Sea, Sea of Okhotsk, coastal waters of the Sea of Japan and Korean Peninsula. The O. ocellatus has been long assigned to the pricklebacks subfamily Opisthocentrinae (Makushok Citation1958; Parin et al. Citation2014). However, following Radchenko (Radchenko Citation2015), based on the comparative molecular phylogenetic analysis of several mitochondrial and nuclear loci, also taking into account morphological traits, the subfamily was raised to family rank (Fricke et al. Citation2018). However, this conclusion is still controversial, the scientific literature and particularly the GenBank Taxonomy Browser still list O. ocellatus in pricklebacks (Stichaeidae) family.
To increase the power of the molecular taxonomy resolution of Opisthocentrus and examine its position within Zoarcales, the mitogenome of O. ocellatus (GenBank accession number MK568985, voucher number MIMB 37599) from Peter the Great Bay (the Vostok Bay; 42.89°N, 132.73°E) of the Sea of Japan collected in May 20, 2017 was determined using dideoxy sequencing technique. Using Sliding Window-Based PSO Algorithm (Yang et al. Citation2011) 27 pars of primers were designed based on the sequences available in GenBank for Zoarcoids. The resulted chromatograms were assembled into mitogenome fragments using Geneious Trial (Kearse et al. Citation2012). Then the produced consensus sequence of complete genome was annotated using MitoAnnotator online pipeline (Iwasaki et al. Citation2013) and MITOS Web Server (Bernt et al. Citation2013).The sequences were aligned by Muscle (Edgar Citation2004) implemented in MEGA 7.0 (Kumar et al. Citation2016) and the TrN + G + I substitution model was chosen as the best for phylogenetic analysis.
The mitogenome of O. ocellatus comprised 16,525 bp in length, including 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and two noncoding regions (CR or D-loop and origin of L-strand replication or OL) which is in accordance with the mitogenomes of the typical ray-fined fish (Kartavtsev et al. Citation2007; Satoh et al. Citation2016). The CR of O. ocellatus located between the tRNAPro and the tRNAPhe genes and is determined to be 850 bp in length. The putative OL is placed in the WANCY region between tRNAAsn and tRNACys with 39 bp in length. Eight tRNAs (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAPro, tRNAGlu) and ND6 are conventionally located on the light strand (L-strand), whereas all of the remaining genes were on the heavy strand (H-strand). The overall base composition of H-strand is 26.6% A, 27.5% T, 18.1% G, and 27.4% C. All 13 genes start with the ATG initiation codon except COI, which initiates with GTG, while ND2 and ND3 terminate with ATA and ATC, respectively. Incomplete stop codons were revealed in ND2, ND3, ND4, COII, ATPase 6, and Cyt-b genes. The atypical stop codons were also found in many other zoacids fishes, such as Stichaeus grigorjewi (Turanov et al. Citation2019), Lycodes brevipes (Li et al. Citation2019), Lycodes ygreknotatus (Zheng et al. Citation2019), X. Atropurpureus (Group et al. Citation2017), and in other taxa (Kartavtsev et al. Citation2007; Satoh et al. Citation2016).
In order to investigate the phylogenetic position of O. ocellatus among nearest relatives, neighbour-joining and maximum likelihood phylogenetic analyses were carried out based on complete mitogenome sequences from 14 fish species of Zoarcales infraorder together with Aulicthys japonicus (Cottioidei, Perciformes) as an outgroup. As shown in , O. ocellatus is more closely related to the family of Pholidae then to Stichaeidae, as well as other families within eelpouts.
Disclosure statement
The authors have no conflicts of interest to declare. The authors are solely responsible for the content and the writing of this paper.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Fricke, R., Eschmeyer, W. N. & Fong, J. D. 2018 ESCHMEYER'S CATALOG OF FISHES: SPECIES BY FAMILY/SUBFAMILY. (http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp). Electronic version accessed 25 December 2018.
- Group HCG, Ayala L, Becerra J, Boo GH, Calderon D, DiMarco A, Garcia A, Gonzales R, Hughey JR, Jimenez GA. 2017. The complete mitochondrial genome of Xiphister atropurpureus (Perciformes: Stichaeidae). Mitochondrial DNA Part B. 2:161–162.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, Nishida M. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Kartavtsev YP, Jung S-O, Lee Y-M, Byeon H-K, Lee J-S. 2007. Complete mitochondrial genome of the bullhead torrent catfish, Liobagrus obesus (Siluriformes, Amblycipididae): genome description and phylogenetic considerations inferred from the Cyt b and 16S rRNA genes. Gene. 396:13–27.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Li Y, Feng J, Song P, Zhang N, Li H, Lin L. 2019. Complete mitochondrial genome characteristics and phylogenetic position of Lycodes brevipes Bean, 1890. Mitochondrial DNA Part B. 4:612–613.
- Makushok VM. 1958. Morfologicheskie osnovy sistemy stikheevykh i blizkikh k nim semeystv ryb (Stichaeoidae, Blennioidei, Pisces)[The morphology and classification of the northern blennioid fishes (Stichaeidae, Blennioidei, Pisces.)]. Tr Zool Instituta Akad Nauk Soiuza Sov Sotsialisticheskikh Resp [Proceedings Zool Inst USSR Acad Sci. 25:3–129.
- Mecklenburg CW, Sheiko BA. 2004. Family Stichaeidae Gill 1864-pricklebacks. Calif Acad Sci Annot Check List Fishes. 35:1–36.
- Parin NV, Evseenko SA, Vasil’eva ED. 2014. Fishes of Russian seas. Moscow: KMK Scientific Press
- Radchenko OA. 2015. The system of the suborder Zoarcoidei (Pisces, Perciformes) as inferred from molecular genetic data. Russ J Genet. 51:1096–1112.
- Satoh TP, Miya M, Mabuchi K, Nishida M. 2016. Structure and variation of the mitochondrial genome of fishes. BMC Genomics. 17:719.
- Turanov SV, Rutenko OA, Kartavtsev YP. 2019. Complete mitochondrial genome of Stichaeus grigorjewi Herzenstein, 1890 (Zoarcales: Stichaeidae). Mitochondrial DNA Part B. 4:899–901.
- Yang C-H, Chang H-W, Ho C-H, Chou Y-C, Chuang L-Y. 2011. Conserved PCR primer set designing for closely-related species to complete mitochondrial genome sequencing using a sliding window-based PSO algorithm. PLoS One. 6:e17729.
- Zheng Y, Chen Y, Chen P. 2019. The complete mitochondrial genome of an eelpout Lycodes ygreknotatus (Teleostei: Zoarcidae). Mitochondrial DNA Part B. 4:616–617.