Abstract
Allium ovalifolium var. leuconeurum is an endemic plant in Southwest China. The complete chloroplast genome sequence of A. ovalifolium var. leuconeurum was generated by de novo assembly using high-throughput sequencing. The length of chloroplast genome was 153,635 bp in length, containing a large-single copy region (LSC) of 82,693 bp, a small-single copy region (SSC) of 17,932 bp, and a pair of inverted repeats (IRa and IRb) regions of 26,505 bp. The genome contains 132 genes, including 86 protein-coding genes, 8 rRNA genes, and 38 tRNA genes. Phylogenetic analysis with the reported chloroplast sequences shows that A. ovalifolium var. leuconeurum is more closely related to Allium prattii, both of which belong to the Sect. Anguinum.
Allium is a large genus of monocotyledonous plants in family Amaryllidaceae (formerly in Liliaceae or Alliaceae) (Chase et al. Citation2009). The genus has a very large variation between species in their genome size (Ricroch et al. Citation2005). Allium ovalifolium Hand.-Mazz. var. leuconeurum J. M. Xu, a perennial herb, is endemic to West Sichuan, China, growing on wet slopes, under ditches or forests at the altitudes of 2500–4000 m. This species belongs to Sect. Anguinum and is characterized by ovate-oblong leaf blade with white main veins, which is of very high ornamental value. Based on our field investigation, the population size of A. ovalifolium var. leuconeurum now is minimal and need urgent conservation. Here, we assembled the complete chloroplast genome of A. ovalifolium var. leuconeurum to provide a gene source for further researches.
Fresh young leaves of A. ovalifolium var. leuconeurum were collected from Lixian county, Sichuan Province, China (N:31°25′57.353″, E:102°44′03.779″). Voucher specimen was deposited in the Herbarium of Beijing Forestry University (BJFC) (under collection numbers of BJFUGWB002). The chloroplast sequence was obtained through the high-throughput sequencing and splicing technology of genomic data (Yu et al. Citation2011). Cetyl trimethylammonium bromide (CTAB) method was used to extract genomic DNAs (Doyle Citation1987) and average 150 bp pair-end sequencing was performed on an Illumina HiSeq 4000 platform (Illumina Inc., San Diego, CA) at Majorbio (http://www.majorbio.com, China). Published chloroplast genome of A. pratti was used as a reference to extract chloroplast reads by Geneious Prime (Kearse et al. Citation2012). Filtered chloroplast reads were exploited for de novo assembly with Geneious Prime (Kearse et al. Citation2012). The chloroplast sequences were annotated by Plann (Huang and Cronk Citation2015), then annotations were verified by Geneious Prime (Kearse et al. Citation2012).
The chloroplast genome of A. ovalifolium var. leuconeurum (Genbank accession no. MK628479) was a circular molecular genome with a size of 153,635 bp in length and had four subregions. 82,693 bp of large-single copy (LSC) and 17,932 bp of small-single copy (SSC) regions are separated by 26,505 bp of inverted repeat (IR) regions. It contained 132 genes (86 protein-coding genes, 8 rRNA genes, and 38 tRNA genes). The overall GC content of the chloroplast genome is 37.0% and those in the LSC, SSC, and IR regions are 34.9, 30.0, and 42.7%, respectively.
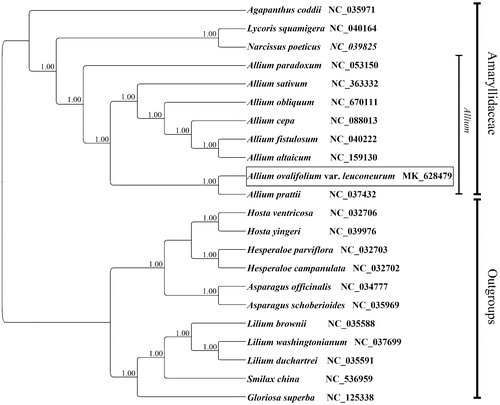
In order to understand the phylogenetic relationship between A. ovalifolium var. leuconeurum and related taxa, the complete chloroplast genome sequences of 10 genera (22 species) were downloaded from the NCBI GenBank database and aligned by MAFFT (Katoh et al. Citation2002). Bayesian inference (BI) was performed with MrBayes version 3.2.3 (Ronquist and Huelsenbeck Citation2003) under the GTR + G+I model to construct the phylogenetic tree. As shows, the posterior probability (PP) of all branch nodes is 1.00. Phylogenetic analysis shows that A. ovalifolium var. leuconeurum is sister to Allium prattii which also belongs to the same Sect. Anguinum (Jing et al. Citation1999). Eight species of Allium are grouped together and are closely related to Lycoris and Narcissus, which supported the present systematic placement of Allium within Amaryllidaceae.
Disclosure statement
The authors declare there is no conflict of interest and are responsible for the content.
Additional information
Funding
References
- Chase MW, Reveal JL, Fay MF. 2009. A subfamilial classification for the expanded asparagalean families Amaryllidaceae, Asparagaceae and Xanthorrhoeaceae. Bot J Linn Soc. 161:132–136.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Jing WC, Xu JM, Yang L. 1999. A study on cytotaxonomy of Sect. Anguinum of Allium. Acta Phytotaxon Sin. 37:20–34.
- Katoh K, Misawa K, Kuma KI, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Ricroch A, Yockteng R, Brown SC, Nadot S. 2005. Evolution of genome size across some cultivated Allium species. Genome. 48:511–520.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Yu HY, Tardivo L, Tam S, Weiner E, Gebreab F, Fan CY, Svrzikapa N, Hirozane- Kishikawa T, Rietman E, Yang XP, et al. 2011. Next-generation sequencing to generate interactome datasets. Nat Methods. 8:478–480.