Abstract
Cercidiphyllum, the only member of Cercidiphyllaceae, contains two extant species: Cercidiphyllum japonicum and Cercidiphyllum magnificum. Here, we reported their complete chloroplast genomes by de novo assembly. The chloroplast genome sizes of C. japonicum and C. magnificum were 159,871 and 159,899 bp, respectively. The two chloroplast genomes have 99.7% sequence identity and were highly conserved in GC content (37.9%), gene content (132 genes), and gene order. Phylogenetic analysis of the chloroplast genomes confirmed the monophyly of Cercidiphyllum and recovered Cercidiphyllaceae as a member of the ‘woody clade’ of Saxifragales.
Cercidiphyllum Sieb. et Zucc., the only genus of Cercidiphyllaceae, contains two extant species Cercidiphyllum japonicum Sieb. et Zucc and Cercidiphyllum magnificum Nakai (Manchester et al. Citation2009). C. japonicum is a long-lived, tall canopy tree occurring in warm-temperate deciduous forests in China and Japan. This species has been treated as ‘endangered’ in China (Fu Citation1992) and recognized as lower risk (LR) by the International Union for Conservation of Nature (IUCN), http://www.iucnredlist.org/). C. magnificum is a small tree or shrub restricted to central Honshu of Japan with smaller number of populations, which is also in urgent need of protection but lacks attention (Chen et al. Citation2010). As cretaceous relict, both species have potential importance in the evolutionary studies of speciation and genetic structuring in Tertiary flora (Qiu et al. Citation2011), which need more comprehensive genetic data. Here, we reported the chloroplast genomes of the two extant species and then performed comparative analyses. In addition, we used the whole chloroplast genomes of two Cercidiphyllum species and 31 related taxa in Saxifragales to reconstructed their phylogenetic relationships.
Samples of C. japonicum and C. magnificum were collected from Mt. Tianmu (Zhejiang, China, 30°19′33.60″N/119°26′31.20″E) and Mt. Tateshina (Nagano, Japan, 36°4″9.10″N/138°17′31.10″E), respectively. Voucher specimens were deposited in the Herbarium of Zhejiang University (HZU). Genomic DNA was extracted from the silica-dried leaves with modified CTAB method (Doyle and Doyle Citation1987). DNA library preparation and 125-bp paired-end sequencing were performed on the Illumina HiSeq2500 platform. The raw data were trimmed and assembled into contigs using CLC Genomics Workbench version 8.5.1 (Qiagen, Aarhus, Denmark). Then all the contigs were aligned with the reference chloroplast genome of Liquidambar formosana (NC_023092; Dong et al. Citation2013) with BLAST (http://blast.ncbi.nlm.nih.gov/) and the draft chloroplast genomes were constructed by connecting overlapping terminal sequences in Geneious version 10.0.5 (http://www.geneious.com). Gene annotation was performed using the Dual Organellar GenoMe Annotator (Wyman et al. Citation2004).
The two chloroplast genomes have a typical quadripartite structure containing a large single-copy region (LSC), a small single-copy region (SSC), and a pair of two inverted repeats (IRs). The chloroplast genome of C. japonicum (MK564061) was 159,871 bp, with 88,023 bp LSC, 18,994 bp SSC, and two 26,427 bp IRs, while C. magnificum (MK550717) was 159,899 bp in genome size, with 88,058 bp LSC, 18,973 bp SSC, and two 26,434 bp IRs. The two chloroplast genomes have 99.7% sequence identity and a total of 111 SNPs and 69 InDels were identified. They were highly conserved in GC content (37.9%), gene order, and gene content (132 genes), including 81 protein-coding genes, 30 tRNA genes, and 4 rRNA genes, of which 17 were duplicated in the IR regions.
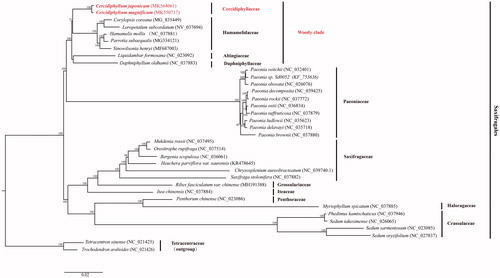
Phylogenetic relationships of two Cercidiphyllum species and other 31 taxa of Saxifragales inferred from maximum likelihood (ML) method using RAxML version 8.2.10 on CIPRES (http://www.phylo.org). The phylogenetic tree () supported the monophyly of Cercidiphyllum and recovered Cercidiphyllaceae as a member of the ‘woody clade’ of Saxifragales (along with Altingiaceae, Daphniphyllaceae, and Hamamelidaceae), consistent with the previous phylogenetic studies (Jian et al. Citation2008; Dong et al. Citation2013, Citation2018).
Disclosure statement
The authors are really grateful to the open raw genome data from public database. No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen C, Liu YH, Fu CX, Qiu YX. 2010. New microsatellite markers for the rare plant Cercidiphyllum japonicum and their utility for Cercidiphyllum magnificum. Am J Bot. 97:e82–e84.
- Dong WP, Xu C, Cheng T, Lin K, Zhou SL. 2013. Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol Evol. 5:989–997.
- Dong WP, Xu C, Wu P, Cheng T, Yu J, Zhou SL, Hong DY. 2018. Resolving the systematic positions of enigmatic taxa: manipulating the chloroplast genome data of Saxifragales. Mol Phylogenet Evol. 126:321–330.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fu LG. 1992. Chinese plant red book. Beijing, China: Science Press.
- Jian SG, Soltis PS, Gitzendanner MA, Moore MJ, Li R, Hendry TA, Qiu YL, Dhingra A, Bell CD, Soltis DE. 2008. Resolving an ancient, rapid radiation in Saxifragales. Syst Biol. 57:38–57.
- Manchester SR, Chen ZD, Lu AM, Uemura K. 2009. Eastern Asian endemic seed plant genera and their paleogeographic history throughout the Northern Hemisphere. J Syst Evol. 47:1–42.
- Qiu YX, Fu CX, Comes HP. 2011. Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol Phylogenet Evol. 59:225–244.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.