Abstract
Disanthus cercidifolius Maximowicz subsp. longipes (H. T. Chang) K. Y. Pan is a rare and Endangered species endemic to central China. Here, we assembled and annotated the complete chloroplast (cp) genome. It is 158,148 bp in length and encodes 87 protein-coding genes, 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. This chloroplast genome sequencing offers a useful resource for future conservation genetics and phylogenetic studies.
Disanthus is a monotypic genus edemic to East Asia. D. cercidifolius subsp. cercidifolius is edemic to Japan, while D. cercidifolius subsp. longipes is endemic to Central China, as a dominant shrub in the mountains at altitudes of 450–1200 m (Zhang et al. Citation2003). This taxon is listed as Endangered by the IUCN (IUCN Citation2017) and rare protected species included in the list of Chinese Dangerous Plant Red Paper (category II) (Fu Citation1992). In the present study, the completed chloroplast genome sequence of D. cercidifolius subsp. longipes is reported contributing for better understanding its evolution and population genetics and also providing significant information for the phylogeny of Hamamelidaceae.
Genomic DNA was extracted from fresh leaves of a seedling of D. cercidifolius subsp. longipes from Pinglang, Duyun, Guizhou, China (107°21′51.92″, 26°02′34.51″ 1163m; Guoxiong Hu et al. 1876, 2018-7-7; KUN), the total genomic DNA was isolated according to a modified CTAB method (Doyle and Doyle Citation1987). Those leaves were stored in the refrigerator at −80 °C. Total genome DNA of D. cercidifolius subsp. longipes was sequenced by Illumina Hiseq 2500 Sequencing System (Illumina, Hayward, CA) to construct the shotgun library and assembled through the NOVOPlasty software (Dierckxsens et al. Citation2017). The low-quality sequences were filtered out Using CLC Genomics Workbench v8.0 (CLC Bio, Aarhus, Denmark) and then reconstructed the chloroplast genome by using MITObim v1.8 (University of Oslo, Oslo, Norway; Kaiseraugst, Switzerland) (Hahn et al. Citation2013), with Sinowilsonia henryi (GenBank: MF687003) as the reference. The complete chloroplast genome of D. cercidifolius subsp. longipes was annotated in Geneious ver. 10.1 (http://www.genei ous.com, Kearse et al. Citation2012) and then submitted to GenBank (accession no. MK411769). The genome annotation was performed by aligning with the cp genomes of relatively related species. Finally, a physical map of the genome was drawn by using the online program OGDRAW (Lohse et al. Citation2013). A voucher specimen of this study is deposited at the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN).
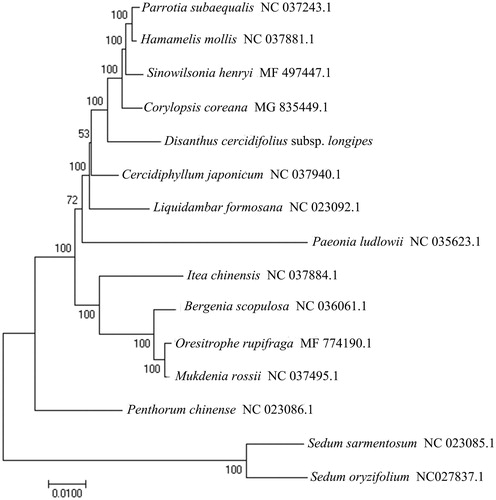
The size of the chloroplast genome of D. cercidifolius subsp. longipes is 158,148 bp, including a large single-copy (LSC) region of 87,138 bp, and a small single-copy (SSC) region of 18,290 bp separated by a pair of identical inverted repeat regions (IRs) of 26,360 bp each. A total of 133 genes were successfully annotated containing 87 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. GC content of the whole genome, IRs, LSC, and SSC regions are 37.9, 43.0, 36.0, and 32.3%, respectively. GC content of IRs region is the highest. Twenty genes contain 1 intron, while 5 genes have 2 introns. The complete chloroplast genome sequence of D. cercidifolius subsp. longipes and other species from Hamamelidaceae were used to construct a phylogenetic tree (). A neighbor-joining (NJ) tree was performed with Mega 6.0 (Tamura et al. Citation2013) using 1000 bootstrap replicates. The result shows the difference of D. cercidifolius subsp. longipes from Saxifragales, which is consistent with previous molecular results (Shi et al.Citation1998; Fishbein et al. Citation2001; Dong et al. Citation2013).
Disclosure statement
The authors declare no conflicts of interest.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Dong W, Xu C, Cheng T, Lin K, Zhou SL. 2013. Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol Evol. 5:989–997.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fishbein M, Hibsch-Jetter C, Soltis DE, Hufford L. 2001. Phylogeny of Saxifragales (Angiosperms, Eudicots): analysis of a rapid, ancient radiation. Syst Biol. 50:817–847.
- Fu LK. 1992. China plant red data book-rare and Endangered plants. Beijing: Science Press.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- IUCN (International Union for Conservation of Nature and Natural Resources). 2017. The IUCN Red List of Threatened species. Version. 2017-2; [accessed 2017 Nov 29]. http://www.iucnredlist.org.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenome- DRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:575–581.
- Shi SH, Chang HT, Chen YQ, Qu LH, Wen J. 1998. Phylogeny of the Hamamelidaceae based on the ITS sequences of nuclear ribosomal DNA[J]. Biochem Syst Ecol. 26:55–69.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zhang Z, Chang HT, Endress PK, 2003. Hamamelidaceae. In: Wu ZY, Raven PH, editors. Flora of China, Vol 9. Beijing: Science Press and Missouri Botanical Garden Press; p. 18–42.