Abstract
Ectomomyrmex javanus Mayr, 1867 is the biggest Ponerine ant in Korean peninsula. We have determined mitochondrial genome of E. javanus. The circular mitogenome of E. javanus is 15,512 bp long including 13 protein-coding genes, 2 ribosomal RNAs, 22 transfer RNAs, and a large non-coding region of 822 bp. The base composition was AT-biased (83.8%). Gene order of E. javanus is identical to Cryptopone sauteri belonging to Ponera genus group which is same to that of E. javanus. Phylogenetic trees show that E. javanus is sister to C. sauteri and both species are outside of the remaining ant mitogenomes agreeing with current phylogeney. The mitochondrial genome of E. javanus will be a milestone for understanding E. javanus in the aspect of molecular phylogeny.
The subfamily Ponerinae is an important ant taxon in that it shows various behavioural traits linked to early evolutions of ant societies (Schmidt and Shattuck Citation2014). Despite its importance, only one Ponerinae mitochondrial genome (Cryptopone sauteri; Park et al. Citation2019) is available, presenting a lack of genomic resources in this subfamily.
Ectomomyrmex javanus Mayr, 1867 is the biggest Ponerine ant in the Korean peninsula. It is identified by sculptured body surface and an oblique furrow on the mesepisternum (Japanese Ant Database Group et al. Citation2003); however, sheer size (over 7 mm) is enough to separate E. javanus from all other Korean Ponerine species. E. javanus shows typical Ponerine traits including monomorphic workers, small colonies made up of a few dozen to above a hundred individuals, and colonies often without dealate queen castes suggesting the existence of gamergates. Nests are made under rocks in forested areas. Workers forage individually on the forest floor hunting for small invertebrates; however, workers occasionally wander on tree trunks during nuptial fights, which is much higher than usual foraging areas.
To understand organelle genome of E. javanus, its total DNA was extracted from workers collected in Mt. Ungilsan, Namyangju-si, Gyeonggi-do, the Republic of Korea using DNeasy Brood &Tissue Kit (QIAGEN, Hilden, Germany). Raw sequences obtained from Illumina HiSeqX at Macrogen Inc., Korea, were filtered by Trimmomatic 0.33 (Bolger et al. Citation2014) and de novo assembled and confirmed by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li et al. Citation2009), and SAMtools 1.9 (Li Citation2013). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used for annotation based on that of C. sauteri (MK138572; Park et al. Citation2019). ARWEN (Laslett and Canbäck Citation2008) was used to annotate tRNAs. The DNA sample and specimen (95% ethanol) are deposited in InfoBoss Cyber Herbarium (IN; Seoul, Republic of Korea; J. Park, KFDS00123).
E. javanus mitochondrial genome length (Genbank accesison is MK496222) is 15,512 bp and its GC ratio is 16.2%, showing an AT-bias. It contains 13 protein-coding genes, 2 rRNAs, and 22 tRNAs. Size of tRNAs ranges from 57 bp to 75 bp, similar to other ants (circa 54–90 bp; Lee et al. Citation2018). Gene order of E. javanus is identical to that of C. sauteri in the same Ponera genus group. Control region is 822 bp (A + T is 89.3%), presumably corresponding to the single largest non-coding AT-rich region.
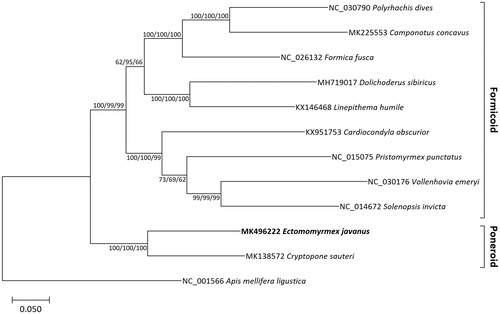
Thirteen protein-coding genes extracted from 11 ant complete mitochondrial genomes including E. javanus and an outgroup species (Apis mellifera ligustica; NC_001566) were aligned by MAFFT 7.388 (Katoh and Standley Citation2013) and then concatnated. Bootstrapped maximum likelihood (bootstrap repeat is 1,000), neighbor joining (bootstrap repeat is 10,000), and minimum evolution (bootstrap repeat is 10,000) trees were constructed using MEGA X (Kumar et al. Citation2018). Phylogenetic trees show that E. javanus is clustered with C. sauteri with high bootstrap values, which is in line with previous phylogeny (Ward Citation2014; ). However, our tree topologies presenting Formicinae is grouped with Dolichoderinae disagree with the previous phylogeny showing Formicinae is grouped with Myrmicinae (Ward Citation2014; ). Ectomomyrmex javanus mitochondrial genome will be a milestone in understanding phylogenetic relationship of Ponerinae species.
Figure 1. Maximum likelihood (bootstrap repeat is 1,000), neighbor-joining (bootstrap repeat is 10,000), and minimum evolution (bootstrap repeat is 10,000) phylogenetic trees of eleven ants and one honeybee mitochondrial genomes: Ectomomyrmex javanus (MK496222 in this study), Cryptopone sauteri (MK138572), Camponotus concavus (MK225553), Polyrhachis dives (NC_030790), Formica fusca (NC_026132), Solenopsis invicta (NC_014672), Cardiocondyla obscurior (KX951753), Vollenhovia emeryi (NC_030176), Pristomyrmex punctatus (NC_015075), Dolichoderus sibiricus (MH719017), Linepithema humile (KX146468), and Apis mellifera ligustica (NC_001566) as an outgroup. Phylogenetic tree was drawn based on maximum likelihood tree. The numbers above branches indicate bootstrap support values of maximum likelihood, neighbor joining, and minimum evolution phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Japanese Ant Database Group, Imai HT, Kihara A, Kondoh M. 2003. Ants of Japan. Japan: Gakken.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 35:1547–1549.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Lee C-C, Wang J, Matsuura K, Yang C-C. 2018. The complete mitochondrial genome of yellow crazy ant, Anoplolepis gracilipes (Hymenoptera: Formicidae). Mitochondrial DNA Part B. 3:622–623.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv:1303.3997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Park J, Kwon W, Park J. 2019. The complete mitochondrial genome of Cryptopone sauteri Wheeler, WM, 1906 (Hymenoptera: Formicidae). Mitochondrial DNA Part B. 4:614–615.
- Schmidt CA, Shattuck SO. 2014. The higher classification of the ant subfamily Ponerinae (Hymenoptera: Formicidae), with a review of ponerine ecology and behavior. Zootaxa. 3817:1–242.
- Ward PS. 2014. The phylogeny and evolution of ants. Ann Rev Ecol, Evol, Syst. 45:23–43.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Gen Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
