Abstract
Gentiana apiata N. E. Br., is a perennial medicinal plant in the family Gentianaceae, which is recorded as near-threatened (NT) species in the Red List and endemic species only distributed in Qinba Mountain, China. The complete chloroplast genome sequence of G. apiata was sequenced using the Illumina Hiseq 4000 platform. The size of the G. apiata chloroplast genome is 151,069 bp, with an average GC content of 37.6%. This circular molecule has a typical quadripartite structure containing a large single-copy (LSC) region of 83,023 bp, a small single-copy (SSC) region of 17,256 bp, and two inverted (IRs) repeat regions of 25,395 bp. A total of 134 genes were successfully annotated containing 86 protein-coding genes, 37 tRNA genes, 8 rRNA genes, and 3 pseudogenes (rps19, ycf1, infA). A maximum likelihood (ML) phylogenetic tree supported the fact that the chloroplast genome of G. apiata is closely related to that of Gentiana hexaphylla.
Gentiana apiata is a perennial herb belonging to genus Gentiana of the family Gentianaceae, commonly known as Tabilongdan or Qinlinglongdan in Chinese medicine (Zhou et al. Citation2009). The genus Gentiana is also a phylogenetic taxon, comprising 361 species; it is the largest genus in the family Gentianaceae (Yuan et al. Citation1996; Wang et al. Citation2017). The alpine plant G. apiata grows only in Qinba Mountains in China, it is an important Chinese folk medicinal plant (Zhou et al. Citation2009). Here, we reported complete chloroplast genomes of G. apiata for the first time based on Illumina Hiseq 4000 pair-end sequencing data.
The plant material of G. apiata was collected from Taibai Mountain, Shaanxi Province, China. The voucher specimen is kept at the Evolutionary Botany Laboratory, College of Life Sciences, Northwest University (Xi’an, Shaanxi, China). Total genomic DNA was extracted from leaf tissue using the Plant Genomic DNA kit (Tiangen Biotech, Beijing, China).
After trimming, the high-quality paired-end reads were assembled with the program MITObim v1.7 (Hahn et al. Citation2013) using the G. macrophylla chloroplast genome sequence as a reference (GenBank accession KY856959) (Wang et al. Citation2017). Annotations were performed using the online program Dual Organellar Genome Annotator (DOGMA) (Wyman et al. Citation2004). We corrected annotations with Geneious (Kearse et al. Citation2012). A map of the genome was generated using OGDRAW (Lohse et al. Citation2013). The chloroplast genome sequence was submitted to GenBank (accession number MK317975).
The chloroplast genome of G. apiata was 151,069 bp in length and contains a pair of inverted repeats (IRa and IRb) regions of 25,395 bp, the large single-copy (LSC) region and small single-copy (SSC) region of 83,023 and 17,256 bp. A total of 134 genes were successfully annotated containing 86 protein-coding genes, 37 transfer RNA genes, 8 ribosomal RNA genes, and 3 pseudogenes (rps19, ycf1, and infA). Among these genes, 15 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpoC1, rps12, rpl16, trnI-GAU, trnA-UGC, trnK-UUU, trnG-UCC, trnL-UAA, and trnV-UAC) have one intron, and three genes (rps12, ycf3, and clpP) have two introns. Overall GC content was 37.6% and in LSC, SSC and IR regions were 35.4%, 31.4% and 43.2%, respectively.
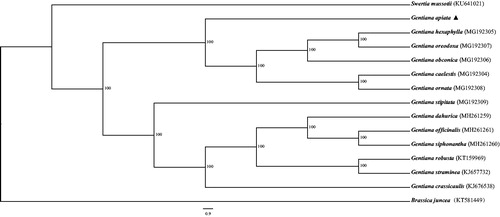
In order to infer the phylogenetic position of G. apiata, we constructed the phylogenetic tree based on the 13 Gentianaceae complete chloroplast genome sequences obtained from NCBI, and Brassica juncea was included in the analysis as an outgroup. All of the chloroplast genome sequences were aligned using MAFFT v7.0.0 (Katoh and Standley Citation2013) and maximum likelihood (ML) analysis was conducted using RAxML (Stamatakis Citation2006). The local bootstrap probability of each branch was calculated by 1000 replications. The resulting tree showed that G. apiata was most closely related to G. hexaphylla with 10 0% bootstrap support (). Above all, this report provides essential data for further study on the accurate identification of species, taxonomy, and phylogenetic resolution of G. apiata, and valuable insight into genetic diversity, conservation, and exploitation efforts for this near threatened (NT) species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Stur-rock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wang X, Yang N, Su J, Zhang H, Cao X. 2017. The complete chloroplast genome of Gentiana macrophylla. Mitochondrial DNA B. 2:395–396.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yuan YM, Kupfer P, Doyle JJ. 1996. Infrageneric phylogeny of the genus Gentiana (Gentianaceae) inferred from nucleotide sequences of the internal transcribed spacers (ITS) of nuclear ribosomal DNA. Am J Bot. 83:641–652.
- Zhou L, Li XK, Miao F, Yang ML, Yang XJ, Sun W, Yang J. 2009. Further studies on the chemical constituents of Chinese folk medicine Gentiana apiata N.E. Br J Asian Nat Prod Res. 11:345–351.