Abstract
Heliciopsis lobata, National class II rare and Endangered plant protection, is one of the subtropical important medicinal plants. However, its complete plastome information and systematic position are still unknown. In order to enhance the development of its Chinese medicinal value and deepen the study of it, we report and characterize the complete plastid genome sequence of H. lobata in an effort to provide genomic resources useful to promoting its medicinal value and systematic research. The complete plastome is 160,961 bp in length and contains the typical structure and gene content of angiosperm plastome, including two inverted repeat (IR) regions of 26,455 bp, a large single-copy (LSC) region of 88,929 bp, and a small single-copy (SSC) region of 19,122 bp. The plastome contains 117 genes, consisting of 83 unique protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes. The overall A/T content in the plastome of H. lobata is 62.00%. The complete plastome sequence of H. lobata will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Proteaceae.
Introduction
Heliciopsis lobata (Merr.) Sleum (Proteaceae, Proteales), National class II rare and Endangered plant protection, is mainly distributed in the southern Guangdong province and Hainan province of China, which has been widely used in the medical and the furniture field. However, its complete plastome information and systematic position is still unknown. In order to enhance the development of its Chinese medicinal value and deepen the study of it, we report and characterize the complete plastid genome sequence of H. lobata in an effort to provide genomic resources useful to promoting its medicinal value and systematic research. H. lobata was sampled from Jianfeng Mountain (18.74°N, 108.86°E), which is a National Nature Reserve of Hainan, China. A voucher specimen (Wang et al., B78) was deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China.
Around 6 Gb clean data were assembled against the plastome of Macadamia integrifolia (KF862711.1) (Nock et al. Citation2014) using MITO bim v1.8 (omicX, France, Hahn et al. Citation2013). The plastome was annotated using Geneious R11.0.4 (Kearse et al. Citation2012) against the plastome of M. integrifolia (KF862711.1). The annotation was corrected with DOGMA (Wyman et al. Citation2004).
The plastome of H. lobata is found to possess a total length 160,961 bp with the typical quadripartite structure of angiosperms, contains two inverted repeats (IRs) of 26,455 bp, a large single-copy (LSC) region of 88,929 bp, and a small single-copy (SSC) region of 19,122 bp. The plastome contains 117 genes, consisting of 83 unique protein-coding genes (seven of which are duplicated in the IR), 30 unique tRNA genes (seven of which are duplicated in the IR) and 4 unique rRNA genes (5S rRNA, 4.5S rRNA, 23S rRNA, and 16S rRNA). Among these genes, 8 pseudogenes (psbA, rps16, rpoC2, rpoB, ycf2, ycf1, ndhA, ycf2), 11 genes possessed a single intron and three genes (ycf3, clpP, rps12) had two introns. The gene rps12 was found to be trans-spliced, as atypical of angiosperms. The overall A/T content in the plastome of H. lobata is 62.00%, which the corresponding value of the LSC, SSC, and IR region were 63.60, 68.50, and 56.90%, respectively.
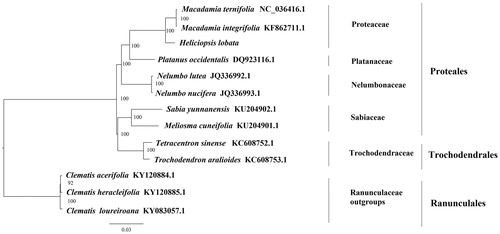
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRCAT substitution model to reconstruct a maximum likelihood (ML) phylogeny of seven published complete plastomes of Proteales, two published complete plastomes of Trochodendrales, using three published complete plastomes of Clematis (Ranunculaceae, Ranunculales) as outgroups. The phylogenetic analysis indicates that H. lobata is more closer to Macadamia integrifolia than other species in this study (). Most of the nodes in the plastome ML tree were strongly supported. The complete plastome sequence of H. lobata will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Proteaceae.
Figure 1. The best ML phylogeny recovered from 13 complete plastome sequences by RAxML. Accession numbers: Heliciopsis lobata (GenBank accession number: MK736992, this study), Macadamia ternifolia NC_036416.1, Macadamia integrifolia KF862711.1, Nelumbo nucifera JQ336993.1, Nelumbo lutea JQ336992.1, Platanus occidentalis DQ923116.1, Sabia yunnanensis, KU204902.1, Meliosma cuneifolia KU204901.1, Trochodendron aralioides KC608753.1, Tetracentron sinense KC608752.1, outgroups: Clematis loureiroana KY083057.1; Clematis heracleifolia KY120885.1; Clematis acerifolia KY120884.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Nock CJ, Baten A, King GJ. 2014. Complete chloroplast genome ofmacadamia integrifoliaconfirms the position of the gondwanan early-diverging eudicot family proteaceae. BMC Genomics. 15:S13.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
