Abstract
The Variegated Racerunner, Eremias vermiculata, is widespread in Northwest China, southern Mongolia, and Zaisan Basin of Kazakhstan. In this study, a nearly complete mitochondrial genome (17,972 bp in length) of E. vermiculata from the Dunhuang Basin in Northwest China was determined by next-generation sequencing. Similar to the typical mtDNA of vertebrates, it contained two ribosomal RNA genes, 13 protein-coding genes (PCGs), 22 transfer RNA genes, and one control region (CR). With exception to the CR, all of the 37 genes were completely recovered. The concatenated PCGs were used to conduct Bayesian phylogenetic analyses together with mitogenome data of lacertids in GenBank. The resulting phylogenetic tree confirmed the monophyly of genus Eremias and its viviparous species as well as E. vermiculata, respectively. The mitogenome presented here will contribute to the examination of phylogeographic structure for E. vermiculata and understanding of mitochondrial DNA evolution in Eremias.
The Variegated Racerunner, Eremias vermiculata, is widespread in Northwest China, southern Mongolia, and Zaisan Basin of Kazakhstan (Szczerbak Citation2003). Despite its wide distribution, this lizard is considered to be a monotypic species without subspecies differentiation (Sindaco and Jeremčenko Citation2008). To date, little has been known regarding its population structure and genetic relationships with other congeneric species (Guo et al. Citation2011).
Recently, three mitochondrial genome sequences of E. vermiculata have been reported (Tong et al. Citation2016; Zhou et al. Citation2016a, Citation2016b). In this study, we determined a nearly complete mitochondrial genome of this species by using next-generation sequencing through Roche 454 GS-FLX technology (Roche, Basel, Switzerland). The lizard was collected from Dunhuang Basin (N39.94°, E94.11°) in July 2013. The Basin is situated on the western end of Northwestern China’s Hexi Corridor and is bounded by the Mingsha, Sanwei, and Beijie Mountains to the southeast, the Mazong and Bei Mountains to the north, and the Kumtag Desert to the west. The specimen (field number Guo2628) was deposited at the herpetological collection in Chengdu Institute of Biology, Chinese Academy of Sciences. We took a similar strategy as described previously (Chen et al. Citation2019) to assemble and annotate the mitogenome by using published sequence (GenBank accession number KP981388) as queries for the reference. A nearly complete mitogenome of 17,972 base pairs (bp) was obtained and deposited in GenBank with accession number MK261078.
The mtDNA sequence comprised two ribosomal RNA genes, 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and one control region (CR or D-loop). The gene content, arrangement, and composition exhibited a typical vertebrate mitochondrial genome feature. The majority of the genes in the mtDNA of E. vermiculata was distributed on H-strand, except for the ND6 gene and 8 tRNAs (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser[UGA], Glu, and Pro). In 13 PCGs, the shortest was ATP8 gene (162 bp) and the longest was ND5 gene (1824 bp). Eleven of 13 PCGs were initiated with the typical ATG codon, except for COI and ND3 with GTG. Most PCGs were terminated with the typical TAA/AGG codons, except for ND2, COII, COIII, ND3, and ND4 with the incomplete termination codon T. The 22 tRNA genes ranged in size from 62 bp for tRNA-Cys to 73 bp for tRNA-Leu and tRNA-Asn. The 12S and 16S rRNA genes were 953 bp and 1530 bp in length, respectively. As for the CR, 1538 bp were already determined adjacent to tRNA-Pro, along with 1047 bp prior to tRNA-Phe.
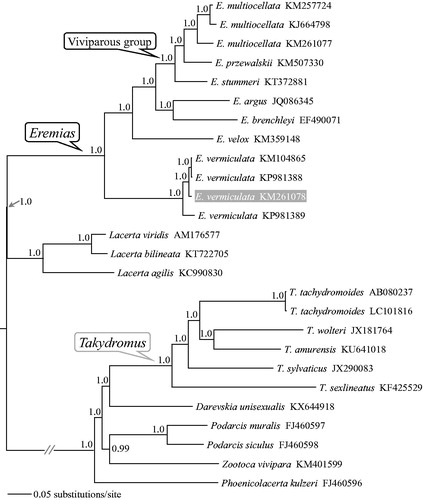
The concatenated PCGs of Eremias available in GenBank plus 14 other lacertids as outgroups were used to reconstruct the Bayesian phylogenetic tree for assessing mitochondrial sequence authenticity of E. vermiculata and its phylogenetic placement. As shown in , the monophyly of both Eremias and the viviparous group were recovered. Four individuals of E. vermiculata clustered together and formed sister taxon to all other sampled members of the genus. The new mitogenome sequence will provide fundamental data for further investigating the phylogeographic structure of E. vermiculata along with detecting mitochondrial DNA evolution in Eremias.
Figure 1. A majority-rule consensus tree derived from Bayesian inference using MrBayes v.3.2.2 (Ronquist et al. Citation2012) with GTR + GAMMA substitution model, based on the concatenated PCGs of 12 individuals of Eremias lizards and 14 outgroups. The tree was rooted using midpoint rooting method and the newly sequenced sample was highlighted in gray. GenBank accession numbers are given with species names. DNA sequences were aligned in MEGA v.7.0.14 (Kumar et al. Citation2016). The PCGs were translated into amino acid sequences and were manually concatenated into a single nucleotide dataset (in total 11,403 bp). Node numbers show Bayesian posterior probabilities. Branch lengths represent means of the posterior distribution.
Disclosure statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Chen D, Li J, Guo X. 2019. Next-generation sequencing yields a nearly complete mitochondrial genome of the Forsyth’s toad-headed agama, Phrynocephalus forsythii (Reptilia, Squamata, Agamidae). Mitochondrial DNA B. 41:817–819.
- Guo X, Dai X, Chen D, Papenfuss TJ, Ananjeva NB, Melnikov DA, Wang Y. 2011. Phylogeny and divergence times of some racerunner lizards (Lacertidae: Eremias) inferred from mitochondrial 16S rRNA gene segments. Mol Phylogenet Evol. 61:400–412.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Sindaco R, Jeremčenko VK. 2008. The reptiles of the Western Palearctic. Vol. 1. Annotated checklist and distributional atlas of the turtles, crocodiles, amphisbaenians, and lizards of Europe, North Africa, Middle East, and Central Asia. Latina, Italy: Edizioni Belvedere.
- Szczerbak NN. 2003. Guide to the reptiles of the Eastern Palearctic. Malabar, Florida: Krieger Publishing Company.
- Tong QL, Yao YT, Lin LH, Ji X. 2016. The complete mitochondrial genome of Eremias vermiculata (Squamata: Lacertidae). Mitochondrial DNA A. 27:1447–1448.
- Zhou T, Liu J, Guo X. 2016a. Complete mitogenome of a Variegated Racerunner, Eremias vermiculata, from north of Tianshan Mountains, Xinjiang, China. Mitochondrial DNA A. 27:2765–2766.
- Zhou T, Wan X, Guo X. 2016b. Sequencing and analysis of the whole mitochondrial genome of a variegated racerunner from Taklamakan Desert. Mitochondrial DNA A. 27:3041–3042.
