Abstract
In this study, the complete mitochondrial genome of Dysomma anguillare was obtained, which was 16,898 bp in length. It consists of 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and a non-coding control region. The overall base composition of the genome is 33.19% for A, 22.9% for T, 28.1% for C, and 15.79% for G. In 13 protein-coding genes, two types of initiation codon (GTG, ATG) and five types of stop codons (AGG,TAA,TAG,TA, and T) were identified. The phylogenetic tree, which is based on complete mtDNA sequences, suggested that D. anguillare was closest to some species of Synaphobranchidae. This study could provide some basic information for further studies on population structure and molecular evolution of D. anguillare.
Dysomma anguillare, an eel in the Order Anguilliformes family Synaphobranchidae, was described by Keppel Harcourt Barnard in 1923 (Barnard Citation1923). It is a marine, tropical fish which is found worldwide in the western Atlantic Ocean and Indo-Western Pacific, including the United States, Venezuela, South Africa, Zanzibar, and Japan. Adults are usually benthic, while larvae are pelagic, mainly at a depth range of 30–270 metres (Bruun Citation1937). In these areas, it is found most commonly at muddy sediments in coastal waters and large rivermouths. Males can reach a maximum total length of 52 centimetres (Fricke and Golani Citation2018). So far, no complete mitochondrial sequence information is available. The study is important for further research on genetics and evolution of D. anguillare.
The specimen of D. anguillare was collected from Fuqing, Fujian, China (25°50′2″N, 119°27′35″E) in April 2017. It was stored in East China Sea Fisheries Research Institute, Chinese Academy of Fishery Science. The genomic DNA was extracted from muscle tissues using Animal Genomic DNA Extraction Kit (Tiangen, Beijing, China) following the operation manual.
We obtained the complete mitochondrial sequence of D. anguillare and submitted it into the Genbank database with an accession number MK574146. It was a circular molecule of 16,898 bp in length, including 13 protein-coding genes, 2 rRNA, 23 tRNA, and a non-coding control region. The overall nucleotide contents of A, T, C, and G are 33.19, 22.9, 28.1, and 15.79 %. In 13 protein-coding genes, two kinds of start codons were identified, CO1 was started with GTG, whereas others started with ATG. Five types of stop codons (AGG, TAA, TAG, TA, and T) were detected and CO1 ended with ATG. Six genes (ND1, ND5, ATP8, ND4L, ND6, and Cytb) ended with TAA or TAG and the others ended with TA or T. Among the 22 tRNAs, 8 tRNAs (tRNAPro, tRNAGlu, tRNASer, tRNATyr, tRNACys, tRNAAsn, tRNAAla, and tRNAGln) were encoded by L-strand, when others were encoded by H-strand. The length of D-loop was 964 bp and its overall nucleotide composition was 36.5% for A, 29% for T, 21.8% for C, and 12.7% for G, with a high AT content of 65.5%.
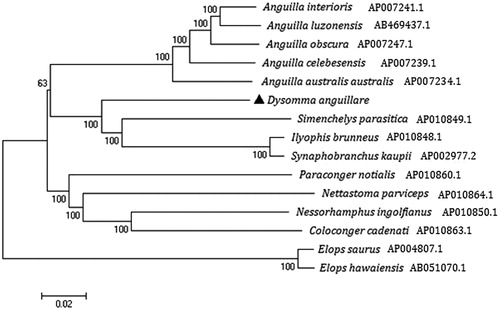
To evaluate the phylogenetic position of D. anguillare, the phylogenetic tree was reconstructed using the neighbor-joining method in MEGA 6 based on complete mtDNA sequences of D. anguillare and other 14 species (Kumar et al. Citation2008) (). Elops saurus (Elopiformes, Elopidae) and Elops hawaiensis (Elopiformes, Elopidae) were used as an out-group. The phylogenetic tree showed that D. anguillare clustered with S.parasitica, I.brunneus, and S.kaupii in family Synaphobranchidae, Then together with some other species in Suborder Anguilloidei and Suborder Congroidei forming a big branch.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Barnard KH. 1923. Diagnoses of new species of marine fishes from South African waters. Annals of the South African Museum. 13:439–445.
- Bruun AF. 1937. Contributions to the life histories of the deep sea eels: Synaphobranchidae. Dana Rep. 9:1–31.
- Fricke R, Golani D. 2018. Dysomma alticorpus, a new species of cutthroat eel from the Gulf of Aqaba, Red Sea (Teleostei: Synaphobranchidae). Comptes Rendus Biolog Ies. 2:111–119.
- Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinfo. 9:299–306.