Abstract
Carpinus laxiflora is a member of the family Betulaceae. In the present study, we analyzed the complete chloroplast genome sequence of C. laxiflora using the Ion Torrent Platform. We found that the chloroplast genome of C. laxiflora was 159,255 bp in length with a large single copy (LSC) region of 88,359 bp, a small single copy (SSC) region of 18,764 bp, and a pair of inverted repeats (IRs) of 26,066 bp. The overall GC content of the chloroplast genome was 36.5%. It contained 135 genes, including 87 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. A phylogenetic analysis indicated that C. laxiflora is closely related to C. monbeigiana.
The genus Carpinus belongs to the family Betulaceae and consists of about 42 species that are mainly distributed between Europe and Eastern Asia (Holstein and Weigend Citation2017). Four of its species (C. cordata Blume, C. turczaninowii Hance, C. tschonoskii Maxim., and C. laxiflora (Siebold and Zucc.) Blume) are distributed in Korea, China, and Japan (Jeon and Chang Citation2000). Carpinus laxiflora is widely used as an industrial tree and timber in Korea (Song and Kim Citation2011). It is a major species of temperate and secondary forests in the temperate zone. Many ecological studies have suggested that C. laxiflora is a representative species of the climax forest in the temperate forests of South Korea (Byeon and Yun Citation2018). Previous studies on C. laxiflora focused on its phylogenetic relationships and were not based on complete chloroplast genome sequencing data (Yoo and Wen Citation2007). In the present study, we characterized the chloroplast DNA from C. laxiflora and confirmed its phylogenetic relationships with another 16 species in Betulaceae.
Individual plant materials were collected from Cheongyang, South Korea (N: 36°24′59.9″, E: 126°51′40.6″). Total DNA was isolated using a Plasmid SV mini kit (GeneAll, Seoul, Korea) and stored in a DNA bank (Division of Forest Genetic Resources DNA bank No.0122099500). Whole genome sequencing data were generated using the Ion Torrent Platform (Life Technologies Corporation, Carlsbad, CA). The sequenced fragments were assembled using Geneious 10.2.3. The tRNAs were confirmed using tRNAscan-SE (Lowe and Eddy Citation1997). A maximum likelihood (ML) tree was conducted using RAxML Blackbox web server, which used the rapid bootstrap method (Stamatakis et al. Citation2008; Stamatakis Citation2014). The tree analysis was conducted using 100 bootstrap replicates.
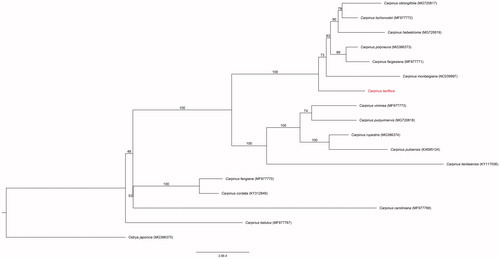
The complete chloroplast genome of C. laxiflora (GenBank: MK425701) was 159,255 bp, with a large single copy (LSC) region of 88,359 bp, a small single copy (SSC) region of 18,764 bp, and 2 inverted repeats (IRa and IRb) of 26,066 bp. The overall GC content was 36.5% (LSC, 34.3%; SSC, 30.1%; IRs, 42.5%). The genome contained 135 genes, including 87 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. Seven of the protein-coding genes, four rRNA genes, and seven tRNA genes were duplicated in the IR regions. Fifteen genes contained a single intron and two genes had two introns (ycf3 and clpP). The results of the phylogenetic analysis determined from the chloroplast genomes of 17 species of Carpinus with an Ostrya species (using the outgroup). The results indicate that C. laxiflora is most closely related to C. monbeigiana (). This information can be used as the basis for a variety of studies using C. laxiflora and population genomic studies of Carpinus species.
Disclosure statement
The authors report no declaration of interest. The authors alone are responsible for the content and writing of this article.
References
- Byeon SY, Yun CW. 2018. Community structure and vegetation succession of Carpinus laxiflora Forest Stands in South Korea. Korean J Environ Ecol. 32:185–202.
- Holstein N, Weigend M. 2017. No taxon left behind? – a critical taxonomic checklist of Carpinus and Ostrya (Coryloideae, Betulaceae). European J Taxon. 375:1–52.
- Jeon JI, Chang CS. 2000. Foliar flavonoids of genus Carpinus in estern Asia primarily based on native taxa to Korea. Korean J Pl Taxon. 30:139–153.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Song KW, Kim YY. 2011. The habitat and utilization of useful plants in Jeju Island. J Educ Res Inst. 13:1–14.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 57:758–771.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Yoo KO, Wen J. 2007. Phylogeny of Carpinus and subfamily Coryloideae (Betulaceae) based on chloroplast and nuclear ribosomal sequence data. Plant Syst Evol. 267:25–35.