Abstract
Debregeasia orientalis is an economically and ecologically important species, which belongs to the genus Debregeasia within the family Urticaceae. The complete chloroplast genome size of D. orientalis is 155,894 bp in length, including a large single copy (LSC) region of 85,558 bp, a small single copy (SSC) region of 19,064 bp, and a pair of inverted repeats (IRs) of 25,636 bp. The genome contains 112 genes, including 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. The GC content in chloroplast genome, LSC region, SSC region, and IR region were 36.3%, 34.0%, 29.4%, and 42.7%, respectively. A total of 15 species are used to construct the phylogenetic tree of Rosales, employing the maximum likelihood (ML), and the results showed that D. orientalis is closely related to Debregeasia saeneb.
Debregeasia orientalis is an economically and ecologically important species in Urticaceae, and it is usually distributed in the shaded, wet places by streams of mountain valleys in East Asia (Chen et al. Citation2003). Previous studies of this species have mainly focused on the chemical constituents (Xiao et al. Citation2008), phylogeny, and biogeography (Wu et al. Citation2013, Citation2015, Citation2018). In this study, we sequenced and assembled the complete chloroplast genome (cpDNA) of D. orientalis, and revealed its closest relative species. The annotated cpDNA of D. orientalis has been deposited into the GenBank with the accession number MH196364.
In this study, young, fresh, and healthy leaves were collected from D. orientalis in Lancang county (Yunnan, China; N 23.238902°, E 99.700676°; Alt. 857m). The voucher specimen was deposited in herbarium KUN (collection numbers is Liuj166488). Total genomic DNA was extracted using CTAB method (Doyle and Doyle Citation1987) and sequenced with Illumina Hiseq 2500 platform. All raw reads were trimmed using NGSQCToolkit_v2.3.3 (Patel and Jain Citation2012). After dislodged the low quality reads, the clean reads were assembled using MIRA v4.0.2 (Chevreux et al. Citation2004) and MITObim v1.8 (Hahn et al. Citation2013) using the chloroplast genome of Morus notabilis (KP939360) as the reference sequences.
The complete chloroplast genome of D. orientalis is 160,621 bp in length, including a large single copy (LSC) region of 88,580 bp, a small single copy (SSC) region of 20,049 bp, and a pair of inverted repeats (IRs) of 25,996 bp (). The GC content in chloroplast genome, LSC region, SSC region, and IR region were 36.2%, 33.9%, 29.0%, and 42.8%, respectively. The complete chloroplast genome of D. orientalis contains 112 genes, including 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. A total of 15 genes (tRNA-Lys(UUU), tRNA-Gly(UCC), tRNA-Leu(UAA), tRNA-Val(UAC), tRNA-Ile(GAU), tRNA-Ala(UGC), rps16, atpF, rpoC1, petB, petD, rps16, rpl2, ndhB, and ndhA) contains a single intron, and three genes (clpP, rps12, and ycf3) contains two introns.
Figure 1. Phylogenetic tree produced by Maximum Likelihood (ML) analysis base on chloroplast genome sequences from 15 species of Rosales, numbers associated with branched are assessed by Maximum Likelihood bootstrap.
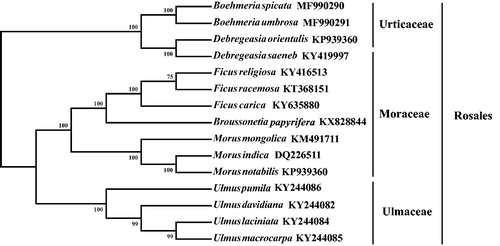
A total of 15 species were used to reconstruct the phylogenetic tree of Rosales (). Modeltest v3.7 (Posada and Crandall Citation1998) was used to determine the best-fitting model. Maximum likelihood analysis was performed using MEGAv7 (Kumar et al. Citation2016) with 1000 bootstrap replicates. The results showed that D. orientalis was closely related to D. saeneb.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14:1147–1159.
- Chen CJ, Lin Q, Friis I, Wilmot-Dear CM, Monro AK. 2003. Urticaceae. In: Wu ZY, Raven PH, editors. Flora of China. Bejing: Science Press, Missouri Botanical Garden Press, p. 76–189.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14:817–818.
- Xiao YH, Cao H, Zhang GL. 2008. Chemical constituents of Debregeasia orientalis. Nat Prod Res & Dev. 20:52–55.
- Wu ZY, Monro AK, Milne RI, Wang H, Yi TS, Liu J, Li DZ. 2013. Molecular phylogeny of the nettle family (Urticaceae) inferred from multiple loci of three genomes and extensive generic sampling. Mol Phylogenet Evol. 69:814–827.
- Wu Z-Y, Milne RI, Chen C-J, Liu J, Wang H, Li D-Z. 2015. Ancestral state reconstruction reveals rampant homoplasy of diagnostic morphological characters in Urticaceae, conflicting with current classification schemes. PLoS One. 10:e0141821.
- Wu Z-Y, Liu J, Provan J, Wang H, Chen C-J, Cadotte MW, Luo Y-H, Amorim BS, Li D-Z, Milne RI. 2018. Testing Darwin's transoceanic dispersal hypothesis for the inland nettle family (Urticaceae). Ecol Lett. 21:1515–1529.
