Abstract
A complete mitochondrial DNA genome of a rare click beetle, Cryptalaus yamato (Nakane), is the first reported. The genome consists of 15,882 base pairs including 13 protein-coding genes (PCGs), 20 tRNAs, two rRNAs, and a 1279 bp long AT-rich region. The overall base composition is 69.4% AT and 30.6% GC. The maximum likelihood analysis based on nucleotide sequence data of 13 PCGs supported that C. yamato is involved in monophyletic group of Elateridae.
The family Elateridae consists of more than 400 genera and almost 10,000 species in all parts of the world (Costa et al. Citation2010). Cryptalaus yamato (Nakane Citation1957), a rare elateridine species which had been regarded as endemic to Japan, was recently recorded in South Korea for the first time with a DNA barcode (COI) sequence by Choi et al. (Citation2019), and any complete mitochondrial genome has not been published in the genus Cryptalaus. In this study, we sequenced the complete mitochondrial genome of C. yamato to accumulate genetic information of elaterid species. A voucher specimen collected from the Gwangneung forest (Pocheon, Gyeonggi province) is deposited in the Korea National Arboretum, South Korea.
The complete mitochondrial genome of C. yamato was sequenced using Illumina MiSeq. A total of 20,201,232 reads were analyzed to generate 3,027,528,810 base pairs of sequence and assembled in Geneious 11.1.5 (Kearse et al. Citation2012). Secondary structure prediction of tRNAs except for tRNAAsn and tRNASer annotated with the improved Mitochondrial Genome annotation (MITOS) webserver (Bernt et al. Citation2013; http://mitos.bioinf.uni-leipzig.de/), and the secondary structures of tRNA genes were analyzed by comparison with the nucleotide sequences of other insect tRNA sequences.
The complete mitochondrial genome of C. yamato is 15,882 bp in length, consisting of 13 protein-coding genes (PCGs), 20 tRNAs, two ribosomal RNAs, and a major non-coding 1278 bp (A + T rich region) and is registered in GenBank (accession number MK524933). The overall base composition is A (40.1%), C (19.7%), G (10.9%), and T (29.3%). A + T content (69.4%) is not much different in that of other elaterids (76.7% in Limonius minutus, 75.3% in Adrastus rachifer, 69.4% in Pyrophorus divergens, 68.6% in Ignelater luminosus, and 63.1% in Pyrearinus termitilluminans). All of the PCGs started with typical ATN codon as the start codon: seven (NAD2, CO2, ATP6, CO3, NAD4, NAD4L, and CYTB) start with ATG and six (CO1, ATP8, NAD3, NAD5, NAD6, and NAD1) with ATT. In addition, the PCGs were terminated with typical TAN codon as the stop codon: eight (NAD2, CO1, CO2, ATP8, ATP6, NADP4, NAD4L, and NAD6) with TAA and five (CO3, NAD3, NAD5, CYTB, and NAD1) with TAG. The A + T rich region of the full-length 1279 bp is located between 12S rRNA and tRNAIle genes. The A + T content of this region contains 71.9%, which is relatively lower than that of other elaterids (87.7% in A. rachifer, 83.2% in L. minutus, 77.5% in I. luminosus, 74.7% in Pyro. divergens, 66.1% in Pyre. termitilluminans).
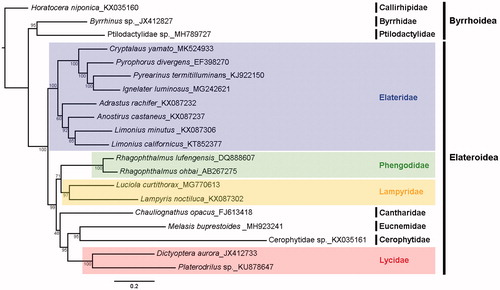
The phylogeny of 17 Elateroidea species and three Byrrhoidea species is reconstructed based on nucleotide sequence data of 13 PCGs with the maximum-likelihood (ML) (). The result well supported that all elaterids are involved in a monophyletic group and this elaterids group is sister to other Elateroidea families. Especially, C. yamato is sister to a clade of Pyrearinus termitilluminans + Ignelater luminosus + Pyrophorus divergens.
Nucleotide sequence accession number
The complete mitochondrial genome sequence of C. yamato has been assigned GenBank accession number MK524933.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Choi I-J, Lee S-G, Suzuki W, Choi S, Lim J. 2019. Taxonomic review of the predatory genus Cryptalaus Ôhira (Coleoptera: Elateridae) from Korea with a new record of C. yamato (Nakane). J Asia Pac Biodivers. https://doi.org/10.1016/j.japb.2019.02.008.
- Costa C, Lawrence JF, Rosa SP. 2010. 4.7. Elateridae Leach, 1815. In: Leschen RAB, Beutel RG, Lawrence JF, editors. Handbook of Zoology, vol 4/39: Coleoptera, beetles, vol. 2. Morphology and systematics (Elateroidea, Bostrichiformia, Cucujiformia partim). Berlin: W Cegruyter; p. 75–103.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desk to software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Nakane T. 1957. New or little-known Coleoptera from Japan and its adjacent regions, XIV. Sci Rep Saikyo Univ. 2:235–240.