Abstract
Cyperus glomeratus plays an important role in sand fixation, soil improvement, and water purification. Here we have sequenced the complete chloroplast genome of the C. glomeratus, which is 167,523 bp in length with large (LSC 81,756 bp) and small single-copy (SSC 9,385 bp) regions separated by a pair of inverted repeats (IRs 38,191 bp) and contains 134 functional genes including 92 protein-coding genes, 8 rRNA genes, and 34 tRNA genes. The phylogenetic analysis showed a strong sister relationship with Hypolytrum nemorum in Cyperaceae. Our findings provide a foundation for further investigation of chloroplast genome evolution in C. glomeratus.
The chloroplast is an organelle unique to green plants and plays an important role in plant growth. As an extranuclear genetic genome, the chloroplast genome encodes proteins involved in photosynthesis and biosynthesis of starch, fatty acids, and other crucial proteins, with the capability of autonomously replicating DNA (Mullet Citation1988). Because the chloroplast genome has the advantages of relatively conservative molecular structure, it is widely used in the fields of phylogeny, molecular evolution, and genetic expression.
Cyperus glomeratus is a pioneer plant of lowland river banks, lakeshores, and is widespread in Europe and Asia (Melečková et al. Citation2016). Cyperus glomeratus was employed in wastewater purification, and phytoremediation for heavy metal contaminated soils (Olson and Fletcher Citation2000; Chang et al. Citation2009). Here, we reconstructed the complete chloroplast genome of the C. glomeratus using high-throughput Illumina sequencing technology, which will help to further study the function of chloroplast genes and its phylogenetic evolution.
DNA samples were extracted from the fresh leaves that were collected from a single individual of C. glomeratus in Xi’an Botanical Garden of Shaanxi Province (Xi’an, Shaanxi, China) and stored in our lab. A total of 31.59 M raw reads were retrieved and trimmed by CLC Genomics Workbench v8.0 (CLC Bio, Aarhus, Denmark). A subset of 20.86 M trimmed reads were used for reconstructing the chloroplast genome by NOVOPlasty (Dierckxsens et al. 2017), with that of its congener Hypolytrum nemorum (GenBank: NC_036036.1) as the initial reference genome. A total of 682,429,193 chloroplast reads yielded an average coverage of 692.6-fold. The chloroplast genome was annotated in GENEIOUS R11 (Biomatters Ltd., Auckland, New Zealand) by aligning with that of H. nemorum.
The chloroplast genome of C. glomeratus is a circular DNA molecule with 167,523 bp in size (MK423990). It comprises a pair of inverted repeat (IR) regions of 38,191 bp each, separated by a large single-copy (LSC) region of 81,756 bp and a small single-copy (SSC) region of 9385 bp. The total GC content is 36.3%, while the corresponding values of the LSC, SSC, and IR regions are 35.2%, 28.7%, and 38.5%, respectively.
This chloroplast genome harbors 134 functional genes, including 92 protein-coding genes (PCGs), 34 tRNA genes and 8 rRNA genes. Among them, 55 are involved in photosynthesis, and 74 genes are involved in self-replication. 63 PCGs and 17 tRNA genes are located in LSC, 6 PCGs and 1 tRNA gene are located in the SSC, while 11 PCGs, 8 tRNA, and all rRNA genes were duplicated in the IR region. Moreover, among all the protein-coding genes, 19 genes contain one intron, while chlp and ycf3 harbor two introns. This is similar to those previously reported for the chloroplast genomes of most other vascular plants (Chumley et al. Citation2006).
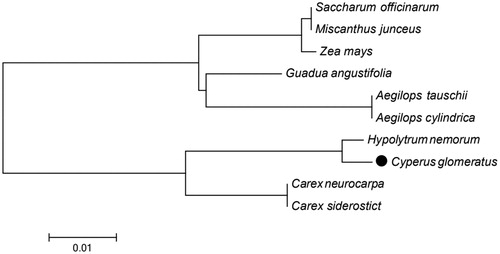
45 photosynthesis related PCGs sequences among 10 chloroplast genomes in Cyperaceae and Gramineae were aligned by MAFFT (Katoh et al. Citation2002) and they were connected as gene strings. The Maximum-Likelihood phylogenetic analysis () showed the position of C. glomeratus was situated as the sister of H. nemorum in Cyperaceae. Our findings provide a foundation for further investigation of chloroplast genome evolution in pioneering plant C. glomeratus.
Figure 1. Phylogenetic tree of 9 species in Cyperaceae and Gramineae based on the Maximum-Likelihood analysis of the whole chloroplast genome sequences using 500 bootstrap replicates. The analyzed species and corresponding Genbank accession numbers are as follows: Aegilops cylindrica (NC_023096), Aegilops tauschii (NC_022133), Carex siderostict (NC_027250), Carex neurocarpa (KU238086), Guadua angustifolia (NC_029749), Hypolytrum nemorum (NC_036036), Miscanthus junceus (NC_035751), Saccharum officinarum (LN849913), Zea mays (NC_001666).
Disclosure statement
The authors report no conflicts of interest, and alone are responsible for the content and writing of the paper.
References
- Chang HQ, Kou TJ, Qiao XH, Jun-Yu HE. 2009. Removal efficiency of nutrients from wastewater by several plants. B Soil Water Conserv. 5:29.
- Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, Jansen RK. 2006. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 1:2175–2190.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 14:3059–3066.
- Lazarević J, Radulović N, Palić R, Zlatković B. 2010. Chemical composition of the essential oil of Cyperus glomeratus L. (Cyperaceae) from Serbia. J Essent Oil Res. 6:578–581.
- Melečková Z, Dítě D, Eliáš P, Schmidt D. 2016. Cyperus glomeratus L.–rediscovered in Slovakia. Hacquetia. 1:93–100.
- Mullet JE. 1988. Chloroplast development and gene expression. Annu Rev Plant Physiol Mol Biol. 1:475–502.
- Olson PE, Fletcher JS. 2000. Ecological recovery of vegetation at a former industrial sludge basin and its implications to phytoremediation. Environ Sci Pollut R. 4:195–204.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Vilhena KSS, Guilhon MSP, Zoghbi MGB, Santos LS, Souza Filho A. 2014. Chemical investigation of Cyperus distans L. and inhibitory activity of scabequinone in seed germination and seedling growth bioassays. Nat Prod Res. 23:2128–2133.
