Abstract
Castanopsis fargesii (Fagaceae) is a major dominant tree species in evergreen broad-leaved forests of subtropical China, with significant ecological and economical values. The complete chloroplast genome sequences of C. fargesii was reported based on the Illumina Hiseq 2500 platform. The complete chloroplast genome was 160,601 bp with a typical quadripartite structure, containing a pair of inverted repeated (IR) regions (25,693 bp) that was separated by a large single copy (LSC) region of 90,258 bp, and a small single copy (SSC) region of 18,987 bp. It harbours 123 functional genes, including 78 protein-coding genes, 37 transfer RNA and 8 ribosomal RNA genes. The ML phylogenetic analysis indicated C. fargesii is closely related to Castanopsis hainanensis.
Castanopsis fargesii Franch. is a major dominant tree species belongs to the genus Castanopsis of Fagaceae family in evergreen broad-leaved tree forests of subtropical China (Sun et al. Citation2014). It is an important ectomycorrhizae tree with significant ecological and economical values, and it is regarded as a potential woody food tree species because of delicious edible fruits (Wang et al. Citation2011). In this study, the complete chloroplast genome sequences of C. fargesii was reported based on the Illumina Hiseq 2500 platform, helpful for well understanding their characteristics and the origin in evolution.
Fresh young leaves of C. fargesii were collected from the Jiulianshan national nature reserve in Ganzhou of Jiangxi province, China (24°33′N, 114°25′E). The voucher specimen was deposited in Jiangxi agricultural university college of forestry in Nanchang, China. Total genomic DNA was extracted using the Plant Genomic DNA Kit, and sequenced with Illumina Hiseq 2500 platform. Approximately 5.88 GB of clean data were yielded. These trimmed reads were assembled by NOVOPlasty (Dierckxsens et al. Citation2017). The assembled genome was annotated using CpGAVAS (Liu et al. Citation2012) and DOGMA (Lohse et al. Citation2007).
The complete chloroplast genome of C. fargesii was 160,601 bp in length with a typical quadripartite structure, containing a pair of inverted repeated (IR) regions (25,693 bp) that was separated by a large single copy (LSC) region of 90,258 bp, and a small single copy (SSC) region of 18,987 bp. The GC content of the whole chloroplast genome was 36.81%. A total of 123 functional genes were annotated, including 78 protein-coding genes, 37 tRNA genes (one for each amino acid, two each for Alanine, Asparagine, Cystine and Valine, three each for Arginine, Glu, Ser and Threonine, four each for Leucine and Methionine, one each for the rest), and 8 rRNA genes. All genes are encoded by 26,583 codons. The annotated chloroplast genome of C. fargesii has been deposited in GenBank with accession number MK571045.
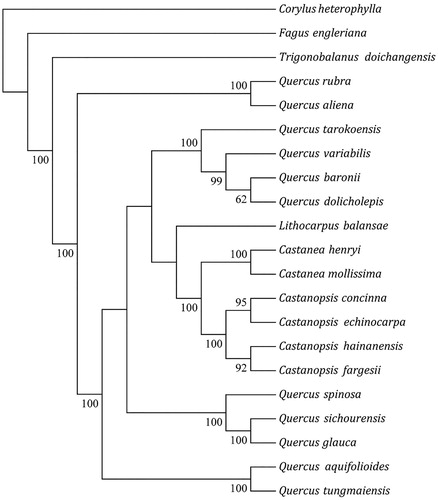
Twenty-one of related chloroplast genome sequences were aligned using MAFFT (Kazutaka et al. Citation2005). Phylogenetic analysis was constructed based on the 21 alignment sequences chloroplast genomes by maximum likelihood (ML) analysis by RaxML based on conducted adopting Kimura 2-parameter model with 1000 bootstrap replicates(Alexandros et al. Citation2017). Corylus heterophylla was used as the outgroup. As shown in the ML phylogenetic tree (), the genus Castanopsis formed a monophyletic clade and that C. fargesii is closely related to Castanopsis hainanensis. These results will lay a basis for the study of phylogeny, phylogeography, and population genetic diversity of C. fargesii.
Figure 1. The ML phylogenetic tree based on 21 complete chloroplast genomes. Accession number: Corylus heterophylla (KX822769), Fagus engleriana (KX852398), Trigonobalanus doichangensis (KF990556), Quercus rubra (JX970937), Quercus aliena (KP301144), Quercus tarokoensis (MF135621), Quercus variabilis (KU240009), Quercus baronii (KT963087), Quercus dolicholepis (KU240010), Lithocarpus balansae (KP299291), Castanea_henryi (KX954615), Castanea mollissima (HQ336406), Castanopsis concinna (KT793041), Castanopsis echinocarpa (KJ001129), Castanopsis hainanensis (MG383644), C. fargesii (MK571045), Quercus spinosa (KM841421), Quercus sichourensis (MF787253), Quercus glauca(KX852399), Quercus aquifolioides (KP340971) and Quercus tungmaiensis (MF593893). The number on each node indicates the bootstrap value.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alexandros S, Paul H, Jacques R. 2017. A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biol. 57:758.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Kazutaka K, Kei-Ichi K, Hiroyuki T, Takashi M. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274.
- Sun Y, Hu H, Huang H, Vargas-Mendoza CF. 2014. Chloroplast diversity and population differentiation of Castanopsis fargesii (Fagaceae): a dominant tree species in evergreen broad-leaved forest of subtropical China. Tree Genet Genom. 10:1531–1539.
- Wang Q, Gao C, Guo L-D. 2011. Ectomycorrhizae associated with Castanopsis fargesii (Fagaceae) in a subtropical forest, China. Mycol Prog. 10:323–332.
