Abstract
Tetraselmis desikacharyi is a marine alga, known as an important plankton for aquaculture as a feed organism. However, the genomic study on this class is rare. Here, we present a complete chloroplast genome of T. desikacharyi, belonging to Chlorodendrophyceae with a full length of 149,934 bp, characterized by a very small single-copy (SSC) region without any genes and a large inverted repeat (IR) region. A maximum-likelihood (ML) phylogenetic analysis was performed using three kinds of data comprising 50 protein-coding genes, which placed the Chlorodendrophyceae as a deep-diverging lineage of the core Chlorophyta.
Tetraselmis desikacharyi, characterized by their intense green colored chloroplast, is important for aquaculture as a feed organism, and for studying plankton growth cycles due to their fast growth rate (Norris et al. Citation1980; Arora et al. Citation2013). However, only two chloroplast genomes have been published in the past years for Chlorodendrophyceae (Turmel et al. Citation2016), which makes the phylogenetic relationship between Chlorodendrophyceae and UTC clade (Ulvophyceae, Trebouxiophyceae, Chlorophyceae) a mystery.
Here, we reported a complete chloroplast genome of T. desikacharyi. The samples were collected from marine envionment at Rochigou, Ile de Batz, Bretagne, France (48°44′43″N, 4°00′35″W). The voucher specimen has been deposited in the Culture Collection of Algae at the University of Cologne (Strain number: CCAC 0023). The algae were grown in Waris-H culture medium (l) (McFadden and Melkonian Citation1986), and the total DNA was extracted using a modified CTAB protocol (Sahu et al. Citation2012). BGISEQ-500 platform was used to generate approximately 50Gb high quality, paired-end (100 bp) reads. CLC-Assembly-Cell-5.1.1 (http://www.clcbio.com/) was used to remove the adapter and unpaired reads. The complete chloroplast genome was assembled by the seed-extension-based de novo assembler NOVOPlasty2.5.9 (Dierckxsens et al. Citation2017), based on an rbcL seed gene sequence of Chlamydomonas reinhardtii. The assembled chloroplast genome was annotated using a web-based program GeSeq (Tillich et al. Citation2017), and the protein-coding genes were further annotated by GeneWise v2.4.1 (Birney and Durbin Citation2000). The plastid genome has been submitted to the CNGB Nucleotide Sequence Archive (CNSA: https://db.cngb.org/cnsa; accession number: CNP0000390).
The complete plastid genome of T. desikacharyi has a total length of 149,934 bp, with a typical quadripartite structure composed of a large single-copy (LSC) region (71,639 bp) and an SSC region (1611 bp) interspersed between the IRa/b region (38,343 bp). It encodes a total of 81 protein-coding genes, six ribosomal RNAs, and 37 transfer RNAs.
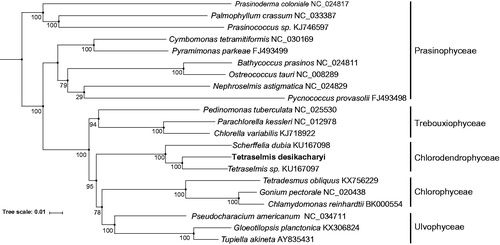
To further investigate its phylogenetic position, a maximum-likelihood phylogenetic tree was generated using RAxML based on complete chloroplast genome sequences along with 20 other Chlorophyta species. Three different data sets were used for the phylogenetic tree construction: (1) all nucleotide positions; (2) the first and second codon nucleotide positions; (3) the amino acid of 50 protein-coding genes. GTRCAT (PROTGAMMAWAG for AA) model was used with 500 bootstraps. The results revealed that the phylogenetic relationship using the three different data sets were highly consistent at the family level, which places the Chlorodendrophyceae as a deep-diverging lineage of the core Chlorophyta ().
Disclosure statement
No potential conflict of interest was reported by the authors
Additional information
Funding
References
- Arora M, Anil AC, Leliaert F, Delany J, Mesbahi E. 2013. Tetraselmis indica (Chlorodendrophyceae, Chlorophyta), a new species isolated from salt pans in Goa, India. Eur J Phycol. 48:61–78.
- Birney E, Durbin R. 2000. Using GeneWise in the Drosophila annotation experiment. Genome Res. 10:547–548.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- McFadden GI, Melkonian M. 1986. Use of Hepes buffer for microalgal culture media and fixation for electron microscopy. Phycologia. 25:551–557.
- Norris RE, Hori T, Chihara M. 1980. Revision of the genus Tetraselmis (Class Prasinophyceae). J Plant Res. 93:317.
- Sahu SK, Thangaraj M, Kathiresan K. 2012. DNA Extraction Protocol for Plants with High Levels of Secondary Metabolites and Polysaccharides without Using Liquid Nitrogen and Phenol. ISRN Mol Biol. 2012:1.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Turmel M, De Cambiaire JC, Otis C, Lemieux C. 2016. Distinctive architecture of the chloroplast genome in the chlorodendrophycean green algae Scherffelia dubia and Tetraselmis sp. CCMP 881. PLoS One. 11:e0148934.