Abstract
Aesculus chinensis Bunge was widely cultivated as a street tree. The complete chloroplast genome of A. chinensis, a plant species with edible seeds in family Sapindaceae, was generated in this study. The length of the complete chloroplast genomes was 155,706 bp and the total GC contents were 38.0%, 85,506 bp of large single copy (LSC) and 18,714 bp of small single copy (SSC) regions were separated by 25,743 bp of inverted repeat (IR) regions. One hundred thirty genes were predicted, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis of A. chinensis and related species revealed that A. chinensis was most closely related to A. wangii.
The genus Aesculus belongs to the family Sapindaceae, Trib. Hippocastaneae. It is known as one of the four world street trees (Wei et al. Citation2008). Aesculus chinensis is also an ornamental garden plant due to large palmate compound leaves and white inflorescence (Wei et al. Citation2008). In many areas, A. chinensis is used as traditional Chinese medicine (Wang and Zhao Citation2001) and has important economic value. Moreover, plants of this genus have a long life span and strong vitality, adapting to various adverse environments and resisting a certain degree of pollution (Lin Citation2010). Few researches on the complete chloroplast genome of Aesculus have been reported. Here, we assembled the complete genome of A. chinensis to provide basic genetic sources for further research.
The mature and healthy leaves were collected from Runcheng town, Yangcheng county, Shanxi province, China (N:35°32′5.442″, E:113°12′39.6″). Voucher specimen was deposited in the Herbarium of Beijing Forestry University (BJFC) (under collection numbers of BJFUGWB007B005). The genomic DNAs were extracted by Plant Genomic DNA Kit. These DNAs were manufactured to average 150 bp paired-end (PE) library and sequenced by Illumina genome analyzer (Hiseq X-Ten, sequencing platform at Yuanyi, Beijing, China, http://www.xn--ori-4n0a Gene.cn/). SOAP de novo was used for preliminary assembly to get contig sequences (Luo et al. Citation2012). The assembled long sequences were then mapped to the closely related species chloroplast genome by using BLAST (Kent Citation2002). The sequences were spliced to obtain a frame map of the chloroplast genome and a circular chloroplast genome sequence was obtained. Cp GAVAS (Liu et al. Citation2012) and DOGMA (Wyman et al. Citation2004) (http://dogma.ccbb.utexas.edu/, China) were used to make a complete annotation. Finally, the annotations were verified by Geneious Prime (Kearse et al. Citation2012).
The chloroplast genome of A. chinensis (Genbank accession no. MK648235) was a circular molecular genome with a size of 155,706 bp in length, 85,506 bp of a large single copy (LSC) and 18,714 bp of small single copy (SSC) regions were separated by 25,743 bp of inverted repeat (IR) regions. It contained 130 genes (85 protein-coding genes, 8 rRNA genes, and 37 tRNA genes). The overall GC content was 38.0% and those in the LSC, SSC, and IR regions were 36.1, 31.8, and 43.2%, respectively.
The phylogenetic tree including Aesculus and other species in Sapindaceae was constructed by complete chloroplast genome sequences of 16 genera (23 species). They were downloaded from the NCBI GenBank database and aligned by MAFFT (Katoh et al. Citation2002). Bayesian inference (BI) was performed with MrBayes v3.2.3 (Ronquist and Huelsenbeck Citation2003) to construct the phylogenetic tree. As shows, the posterior probability (PP) of all branch nodes proves high credibility of the tree.
Figure 1. Phylogenetic relationships of 23 species based on chloroplast genome sequences. PP values for Bayesian analysis are shown at each node.
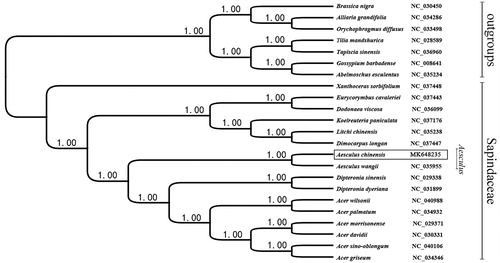
The phylogenetic analysis showed that A. chinensis was closely related to A. wangii (). In addition, all clades in Xanthoceratoideae, Hippocastaneae, Aceroideae, and Dodonaeoideae were supported by high bootstrap values, agreeing with previous studies (Chase et al. Citation2016). The complete chloroplast genome of Aesculus would be used for population genomic studies, phylogenetic analyses, and genetic engineering studies to contribute to our better development and utilization of this species.
Disclosure statement
The authors declare there are no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Chase MW, Christenhusz MJM, Fay MF, Byng JW, Judd WS, Soltis DE, Mabberley DJ, Sennikov AN, Soltis PS, Stevens PF. 2016. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Katoh K, Misawa K, Kuma KI, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kent WJ. 2002. BLAT-the BLAST-like alignment tool. Genome Res. 12:656–664.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. Cp GAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Lin HY. 2010. Biological characteristics and application development of Aesculus chinensis. Anhui Agric Sci Bull. 16:92–93. (Chinese).
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, et al. 2012. SOAP de novo 2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Wang XY, Zhao YF. 2001. Studies on chemistry ingredients of Aescali and medical-value of Aescin. Tangshan Teach Coll. 5:7–11. (Chinese).
- Wei JX, Jiang WB, Weng ML. 2008. Cultural connotations of Aesculus chinensis and their application in landscape architecure. Chin Agric Bull. 24:356–359. (Chinese).
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
