Abstract
Cryptotaenia japonica Hassk. (Apiaceae) is a herbaceous species naturally distributed in China. The complete chloroplast genome sequence of C. japonica was generated by de novo assembly using whole genome next generation sequencing data. The chloroplast genome of C. japonica was 153,259 bp in length and divided into four distinct regions, such as large single copy region (83,462 bp), small single copy region (16,968 bp) and a pair of inverted repeat regions (26,425 bp and 26,406 bp). The genome annotation predicted a total of 133 genes, including 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Phylogenetic analysis with the reported chloroplast genomes revealed that C. japonica is nested in Apiaceae tribe Oenantheae and has close relationship to Cicuta virosa and Tiedemannia filiformis subsp. Greenmannii.
Keywords:
Cryptotaenia japonica Hassk. (Apiaceae) is an herbaceous plant species naturally distributed in China. It wildly grows in forests and ditches, 200–2400 m, and is used in traditional Chinese medicine as a tonic for building the body. It is a distinctive and widespread taxon showing almost continuous variation in leaf and inflorescence form across the range. It is here treated as a species closely resembling, but distinct from, the North American Cryptotaenia canadensis (Linnaeus) de Candolle (Pan and Watson, Citation2005). Although several phylogenetic studies reported nuclear and chloroplast sequences of C. japonica (Kondo et al. Citation1996; Gong et al. Citation2018), the complete chloroplast genome sequence is not available till now. Here, we report the complete chloroplast genome sequence of C. japonica to provide a genomic resource and to clarify phylogenetic relationship of this plant with other species in the Apiaceae family. Total genomic DNA was isolated from mature leaves sampled from the campus of Sichuan University, Chengdu, Sichuan, China. Voucher specimens were deposited in SZ (Sichuan University Herbarium). Chloroplast DNA (cpDNA) was extracted using High-salt Low-pH (HSLp) method and Sucrose DNase (SucDNase) method. The isolated genomic was manufactured to average 400 bp paired-end (PE) library using Illumina Hiseq platform (Illumina, San Diego CA), and sequenced by Illumina genome analyser (Hiseq PE150). The raw reads were then assembled using NOVOPlasty 2.7.2 (Dierckxsens et al. Citation2017) with ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcL) gene from Cicuta virosa as seed. The draft sequence obtained from NOVOPlasty was corrected manually by clean read mapping using bowtie2 (Langmead and Salzberg Citation2012) and Tablet (Milne et al. Citation2013). The genes in chloroplast genome were predicted using Geneious (Kearse et al. Citation2012) and corrected manually. The complete chloroplast genome of C. japonica (GenBank accession no. MK629764) was a circular form of 153,259 bp in length, which was separated into four distinct regions such as large single copy (LSC) region of 83,462 bp, small single copy (SSC) region of 16,968 bp and a pair of inverted repeat regions of 26,425 bp and 26,406 bp. Overall GC contents of chloroplast genomes were 37.5%. The chloroplast genome contained a total of 133 genes including 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes.
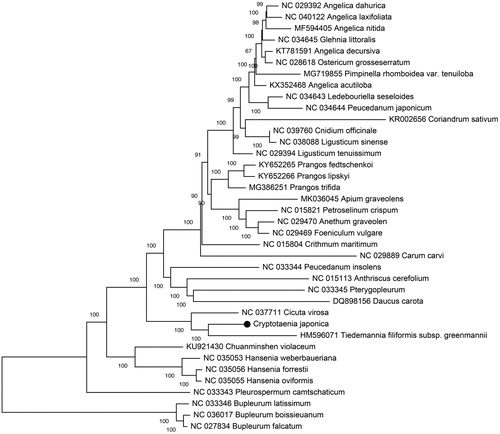
In order to understand the phylogenetic relationship between C. japonica and related species, the complete chloroplast genome sequences of 27 genera (40 species) from Apiaceae were aligned using MAFFT (Katoh et al. Citation2002) and trimmed properly using trimAl v1.4 (Capella-Gutierrez et al. Citation2009). The evolutionary history was inferred using the Neighbour-Joining method in MEGA7.0 (Kumar et al. Citation2016). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap (BS) test (100,000 replicates) are shown next to the branches (). As was expected, C. japonica was placed within Apiaceae tribe Oenantheae, and comprise a clade with Cicuta virosa and Tiedemannia filiformis subsp. greenmannii with 100% BS value.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Pan FD, Watson MF. 2005. Cryptotaenia de Candolle In: Flora of China Editorial Committee, editor. Flora of China, vol 14. St. Louis: Missouri Botanical Garden Press; p. 80
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25:1972–1973.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18
- Gong L, Qiu XH, Huang J, Xu W, Bai JQ, Zhang J, Su H, Xu CM, Huang ZH. 2018. Constructing a DNA barcode reference library for southern herbs in China: a resource for authentication of southern Chinese medicine. PLoS ONE. 13: e0201240
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kondo K, Terabayashi S, Okada M, Yuan C, He S. 1996. Phylogenetic relationship of medicinally important Cnidium offcinale and Japanese apiaceae besed on rbcL sequences. J. Plant Res. 109:21–27.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–354.
- Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 14:193–202.