Abstract
We sequenced and characterized the complete mitochondrial genome of the Madagascan plowshare tortoise Astrochelys yniphora (Testudines, Testudinidae). This genome was 16,527 bp and consisted of 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and a noncoding control region. The genome organization, gene order, and base composition were identical to the typical vertebrate. There was no an additional C in arm of tRNALys gene secondary structure. Astrochelys yniphora 945 bp control region concluded the variable number of tandem repeats (VNTRs). The complete mitogenome of A. yniphora provides the basic data to molecular systematics research of Testudinidae.
The critically endangered plowshare tortoise (Astrochelys yniphora) is endemic to Madagascar and is one of the rarest tortoises (Morgan and Chng Citation2018). Astrochelys yniphora belongs to the genus Astrochelys within the family Testudinidae. At present, only partial mitochondrial genome of A. yniphora has been published on NCBI (Reid et al. Citation2011), the complete mitochondrial genome sequence has not been reported.
The specimen of A. yniphora (Code No. 26080218) was collected from Shanghai Zoo of China (CN) (32°42′N, 122°52′E) and stored in Conservation and Exploitation Research of Biological Resources Lab of Anhui Normal University. Total genomic DNA was extracted from the muscle tissues by the phenol/chloroform method (Zhou et al. Citation2015). The mitochondrial sequence was amplified by PCR using sixteen pairs of primers and recovered by Gel Extract Purification Kit (TaKaRa, Dalian). Purified PCR products were cloned and sequenced with ABI3730 automated sequencer (Invitrogen Co., Ltd, USA). BioEdit v7.2.3 was used to sequence assembly after sequencing (Xiong et al. Citation2010). Protein-coding genes were then aligned using TranslatorX (Abascal et al. Citation2010). The tRNA gene was localized with tRNAscan-SE v2.0, the rRNA gene and control region were aligned with other turtles (Li et al. Citation2017). The complete mitochondrial genome sequence of A. yniphora was submitted to GenBank (accession number JX317746.1).
The A. yniphora complete mitochondrial genome was a circular molecule of 16,527 bp. The arrangement of mitogenes was in agreement with the general features of vertebrates and consists of two rRNA genes, 22 tRNA genes, 13 protein-coding genes (PCGs), and one control region (Liu et al. Citation2018). Total contents of T, C, A, and G were 25.8%, 26.6%, 35.1%, and 12.6%, respectively. The results showed that the anti-G bias of mtDNA based on low content of G which conserved with other vertebrates. The lengths of 16S rRNA and 12S rRNA genes were 1607 bp and 973 bp, respectively. There was no an additional C in arm of tRNALys gene secondary structure, this feature as an identification feature of the mitochondrial genome of Testudinidae, which consistent with previous reports. The length of control region was 945 bp, the variable number of tandem repeats (VNTRs) concluded motifs CAATA and CAATT were identified in this region.
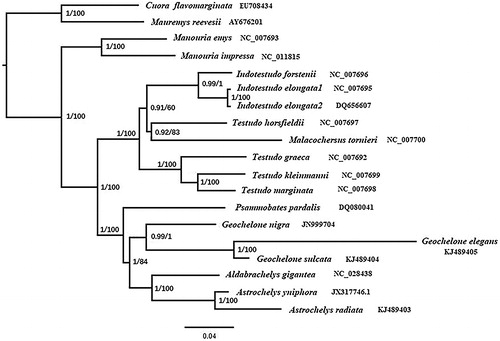
Phylogenetic trees obtained using the Bayesian and maximum-likelihood (ML) methods were depicted in , based on 13 PCGs datasets of 17 Testudinidae species and two Geoemydidea species with RAxML v8.2.12 (Stamatakis Citation2014) and MrBayes v3.2.7 (Huelsenbeck and Ronquist Citation2001), two Geoemydidea species as outgroups. The result indicated a sister relationship between A. yniphora and A. radiata, A. yniphora lies within the genus Astrochelys and has a sister group relationship with Aldabrachelys, combined with the genus Centrochelys forming a monophyletic clade, and establishing a sister group relationship with the genus Stigmochelys. The evolutionary relationships of these species were consistent with the previously reports (Fritz and Bininda-Emonds Citation2007; Reid et al. Citation2011; Lourenco et al. Citation2012), but the phylogenetic positions of these remains to be further studied.
Acknowledgements
The authors are grateful for the comments provided by the reviewers and editors regarding the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–W13.
- Fritz U, Bininda-Emonds O. 2007. When genes meet nomenclature: tortoise phylogeny and the shifting generic concepts of Testudo and Geochelone. Zoology. 110:298–307.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Li H, Liu J, Xiong L, Zhang H, Zhou H, Yin H, Jing W, Li J, Shi Q, Wang Y, et al. 2017. Phylogenetic relationships and divergence dates of softshell turtles (Testudines: Trionychidae) inferred from complete mitochondrial genomes. J Evol Biol. 30:1011–1023.
- Liu J, Jiang H, Zan J, Bao Y, Dong J, Xiong L, Nie L. 2018. Single-molecule long-read transcriptome profiling of Platysternon megacephalum mitochondrial genome with gene rearrangement and control region duplication. RNA Biol. 15:1244–1249.
- Lourenco JM, Claude J, Galtier N, Chiari Y. 2012. Dating cryptodiran nodes: origin and diversification of the turtle superfamily Testudinoidea. Mol Phylogenet Evol. 62:496–507.
- Morgan J, Chng S. 2018. Rising internet-based trade in the Critically Endangered ploughshare tortoise Astrochelys yniphora in Indonesia highlights need for improved enforcement of CITES. Oryx. 52:744–750.
- Reid BN, Le M, McCord WP, Iverson JB, Georges A, Bergmann T, Amato G, Desalle R, Naro-Maciel E. 2011. Comparing and combining distance-based and character-based approaches for barcoding turtles. Mol Ecol Resour. 11:956–967.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Xiong L, Nie L, Li X, Liu X. 2010. Comparison research and phylogenetic implications of mitochondrial control regions in four soft-shelled turtles of Trionychia (Reptilia, Testudinata). Genes Genomics. 32:291–298.
- Zhou H, Jiang Y, Nie L, Yin H, Li H, Dong X, Zhao F, Zhang H, Pu Y, Huang Z, et al. 2015. The historical speciation of Mauremys Sensu Lato: ancestral area reconstruction and interspecific gene flow level assessment provide new insights. PLoS One. 10:e0144711