Abstract
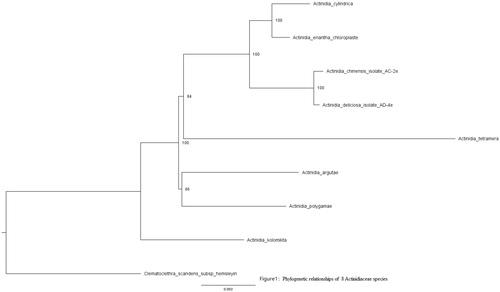
The complete chloroplast (cp) genome sequence of Actinidia cylindrica C. F. Liang was assembled by Illumina paired-end sequencing data in this study. The complete chloroplast genome of Actinidia cylindrica C. F. Liang is 224,773 bp in length, including a large single copy region (LSC) of 158,954 bp, a small single copy region (SSC) of 41,899 bp, and two 11,960 bp inverted repeat regions (IR). A total of 131 genes were annotated, containing 84 protein-coding genes, 39 tRNA genes, 8 ribosomal RNA genes. The phylogenetic position based on the chloroplast genome showed that Actinidia cylindrica C. F. Liang was sister to the group Actinidia eriantha.
Actinidia cylindrica C. F. Liang is an indigenous species in Guangxi, China. Its fruit is tasty. This species is drought-tolerant. Its important commercial value is that the fruits are rich in nutrients, especially high levels of essential minerals, fibers, and vitamin C that are good for the human body. Actinidia cylindrica C. F. Liang has nutritional constituents, including 100 mg/100 g Vc, 7% soluble solid, 1.1% acid, 4.45% total sugar, 6.61% amino acids (Huang et al. Citation2000). In this study, we reported the complete chloroplast genome of Actinidia cylindrica C. F. Liang based on Illumina pair-end sequencing data and revealed its phylogenetic relationship with seven Actinidiaceae species.
We collected fresh leaves from a single individual of this species from Rongshui (N25°07′, E109°13′) and dried them immediately by silica gels. Specimens of Actinidia cylindrica C. F. Liang were stored at a herbarium ar the Institute of Botany, Jiangsu Province and Chinese Academy of Sciences (NAS). Qualified genomic DNA was extracted from young leaves. The improved CTAB method was used to prepare the kiwi fruit genomic DNA. Paired-end and mate-pair Illumina genomic DNA libraries were constructed. The whole-genome sequencing was finished with 150 bp pair-end reads using the Illumina Hiseq Platform. PCR duplicates, adaptor sequences, and low-quality reads were removed. The qualified cleaned reads were assembled into two contigs using NOVOPlasty (Dierckxsens et al. Citation2017). Gaps between two contigs were filled with Geneious (Kearse et al. Citation2012). The genome was annotated using Plann (Huang and Cronk Citation2015). The complete chloroplast genome sequence together with gene annotations were submitted to GenBank under the accession number MK550716.
The complete chloroplast genome of Actinidia cylindrica C. F. Liang is 224,773 bp in length, including a large single copy region (LSC) of 158,954 bp and a small single copy region (SSC) of 41,899 bp, and two 11,960 bp inverted repeat regions (IR). A total of 131 genes are annotated, containing 84 protein coding genes, 39 tRNA genes, 8 ribosomal RNA genes. Nineteen genes are of 2 copies, including 3 protein coding genes(ycf15,ycf2,ndhB), 6 rRNA genes (rps12, rps7,rrn16,rrn23,rrn4.5,rrn5),9 tRNA genes (trnA-UGC,trnH-GUG,trnI-CAU,trnI-GAU,trnL-CAA,trnN-GUU, trnR-ACG, trnV-GAC, trnfM-CAU). The GC-content of the whole chloroplast genome is 37.2%.
Eight Actinidiaceae chloroplast genomes were aligned using MAFFT (Katoh and Standley Citation2013). The phylogenetic tree was built using RaxML (Stamatakis Citation2014) based on neighbor-joining method. The parameters used in the tree construction were gamma models and 1000 bootstraps. A phylogenetic tree was displayed using FigTree7 (CitationRambaut). Confidence ratios of the phylogenetic tree are above 80%.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Figure 1. Phylogenetic relationships of eight Actinidiaceae species based on chloroplast sequences. Bootstrap percentages are indicated for each branch. GenBank accession numbers: Actinidia deliciosa (NC_026691.1), Actinidia kolomikta (NC_034915.1), Actinidia chinensis (NC_026690.1), Actinidia polygama (NC_031186.1), Actinidia eriantha(NC_034914.1), Actinidia tetramera (NC_031187.1), Actinidia arguta (NC_034913.1), and Clematoclethra scandens subsp. hemsleyi (KX345299.1).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Huang H, Gong J, Wang S, He Z, Zhang Z, Li J. 2000. Genetic diversity in the genus Actinidia. Chin Biodivers. 1:1–12.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Rambaut website. [accessed 2015 Feb]. http://tree.bio.ed.ac.uk/software/figtree.
