Abstract
In this study, the complete mitochondrial genome of Pagetopsis macropterus was obtained, which was 17,342 bp including 2 ribosomal RNAs, 13 protein-coding genes, 22 transfer RNAs, and one control region. Of the 37 genes, 28 were encoded on the heavy strand, while 9 were encoded on light strand. The overall nucleotide composition of mitogenome is 27.08% for A, 25.75% for T, 29.78% for C, 17.39% for G, respectively, in accordance with structural characteristics of AT rich. Further, the phylogenetic tree based on complete mtDNA sequences revealed that the P. macropterus was closest to some species of Channichthyidae. This study could provide some valuable information for the studies on evolution for low-temperature adaptability, stock evaluation, and population genetics of P. macropterus.
Pagetopsis macropterus, an important Antarctic fish species of family Channichthyidae, is widely distributed in the Circum-Antarctic on the continental shelf and South Shetland Islands. It constitutes Pagetopsis genus with Pagetopsis maculatus. In these areas, it lives in the depth of 5–655 m of water and it is found most commonly at depths from 5 to 40 m. Their maximum length is 33 cm (Mesa et al. Citation2004). They are the predators in the Antarctic food web. P. macropterus mainly preys on Antarctic krill and some fishes (Artigues et al. Citation2003). So far, no complete mitochondrial sequence information is available. The study is important for the Antarctic ecosystem and further research on genetics and evolution of P. macropterus.
Adult fish of P. macropterus was collected from the Antarctic (58°60′17″W, 62°93′17″S). It was transported to East China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences after freezing at −80 °C. The genomic DNA was extracted from muscle tissue using Animal Genomic DNA Extraction Kit (TIANGEN, Beijing, China) according to the operation manual. In this study, we obtained the complete mitochondrial genome sequence of P. macropterus and submitted it into the GenBank database with an accession number MK517525. This mitochondrial genome is 17,342 bp in length and including 13 protein-coding genes, 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 control region. Among the 37 genes, 28 were encoded on the heavy strand, while 9 were encoded on light strand just as in other teleosts. The overall base composition of this genome is 27.08% for A, 25.75% for T, 29.78% for C, 17.39% for G. The A + T content (52.83%) is similar to Chionodraco hamatus (Song et al. Citation2016), in accordance with the structural characteristics of AT rich. Three kinds of start codons (ATG, GTG, ATC) were identified in 13 protein-coding genes and the rare start codon ATC started ND4. Four types of stop codons (TAA, TAG, TA, T) were detected, seven genes (ND1, ND2, COX1, ATP6, COX3, ND4L, ND6) ended with TAA, five genes (COX2, ND3, ND4, ND5, CYTB) had incomplete stop codons TA or T, one gene (ATP8) ended with TAG, it was different from Electrona carlsbergi (Liang et al. Citation2018). The putative control region (D-loop) was 1144 bp in length, which has a higher A + T content (61.23%) is 712 bp and its overall nucleotide composition is 33.30% for A, 28.93% for T, 21.50% for sCand16.26% for G.
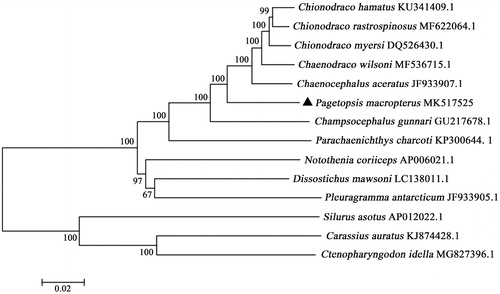
To assess its phylogeny and evolution, the phylogenetic tree was reconstructed based on complete mtDNA sequences using the neighbor-joining method in MEGA 5.1 (Kumar et al. Citation2008) (). Silurus asotus, Carassius auratus, and Ctenopharyngodon idella were used as an out-group. The NJ tree showed that P.macropterus clustered with other species in family Channichthyidae, forming a big branch. Besides, three species in family Nototheniidae formed a big sister branch as well.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Artigues B, Morales-Nin B. Balguerías E. 2003. Fish length-weight relationships in the Weddell Sea and Bransfield Strait. Polar Biol. 26: 463–467.
- Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinformatics. 9:299–306.
- Liang SZ, Song W, Ma CY, Zhang FY, Jiang KJ, Wang LM, Ma LB. 2018. The complete mitochondrial genome of Electrona carlsbergi (Myctophiformes, Myctophidae) with phylogenetic consideration. Mitochondr DNA B. 3: 151–152.
- Mesa ML, Eastman JT, Vacchi M. 2004. The role of notothenioid fish in the food web of the Ross Sea shelf waters: a review. Polar Biol. 27: 321–338.
- Song W, Li LZ, Huang HL, Meng YY, Jiang KJ, Zhang FY, Chen XZ, Ma LB. 2016. The complete mitochondrial genome of Chionodraco hamatus (Notothenioidei: Channichthyidae) with phylogenetic consideration. Mitochondr DNA B. 1:52–53.