Abstract
The complete mitochondrial genome of the Apogonichthys lineatus has been determined in this study. The complete genome (mtDNA) is 16,510 base pairs in length and contains 13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes. The overall base composition of the genome in descending order was28.4% C, 27.3% A, 26.7% T, 17.6% G. In 13 protein-coding genes, most of the protein-coding genes start with ATG, except ND1 start with ATA, COX3 and ND5 start with ATT. For the stop codon, nine protein-coding genes end with TAA, two protein-coding genes end with TAG (ND3 and ND1) and two protein-coding genes end with TGA (ND4 and COX2). Phylogenetic analysis indicated that A. lineatus is close to Pterapogon kauderni families. This study was determined the complete mitochondrial genome of Apogonidae species. It would be a supplement for the genetic analysis of A. lineatus and promote the phylogenetic of Apogonidae.
Apogonichthys lineatus belongs to the family of Apogonidae (Gehrke Citation1988). It widely distributes in the Western North Pacific. It is a common species that can be found in the north of the East China Sea, the Yellow Sea, Bohai Sea (Lin et al. Citation2009). The length of the adult fish is between 40 mm and 60 mm. It’s a small fish species and its spawning period is from August to September. The A. lineatu is a kind of important bait fish which is the dominant species of small fishes and carnivorous economic fishes in the East China Sea area (Zhang et al. Citation2005). Due to its small size and no direct use value, like other small fish species, it has been neglected (Li et al. Citation2004). The otoliths of small A. lineatu are oval in shape and broad in the back. In the previous studies, otolith morphology was identified automatically, but it could not accurately determine its shape (Gehrke Citation1987).
Here, it is the first study which determined the complete mitochondrial genome of A. lineatus. The specimen of A. lineatus was collected from Zhejiang province, China (122.2°E, 30.3°N) and identified by morphology and deposited in the National Engineering Research Center for Marine Aquaculture, Marine Science and Technology College, Zhejiang Ocean University. The total DNA extraction was utilized using the salting-out method with the muscle. The Illumina HiSeq X Ten platform (Shanghai, China) was used to perform the high-throughput sequence.
The complete mitochondrial genome of A. lineatus is 16,510 bp in length (GenBank accession number: MK537324). The complete mitochondrial genome has 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes (tRNA). The nucleotide composition for A. lineatus is 27.3% A, 26.7% T, 28.4% C, and 17.6% G. In 13 protein-coding genes, most of the protein-coding genes start with ATG, ND1 start with ATA, COX3, and ND5 start with ATT. For the stop codon, nine protein-coding genes end with TAA, two protein-coding genes end with TAG (ND3 and ND1) and two protein-coding genes end with TGA (ND 4 and COX2). The 16S rRNA is 1679 bp between the tRNAVal and tRNALeu2 and the 12S rRNA is 955 bp between the tRNAPhe and tRNAVal.
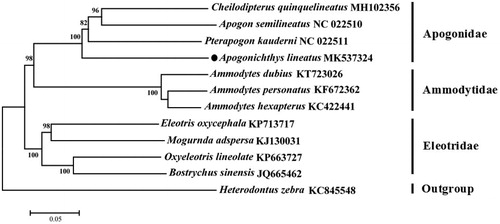
The phylogenetic tree () was constructed based on 13 protein-coding genes of 12 species using the neighbor-joining method in the program Phylip (Felsenstein Citation1993). The result showed that A. lineatus is close to Pterapogon kauderni families. We believe that this result will further supplement the genome information in mitochondria of the family Apogonidae and facilitate the study on population genetic.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Felsenstein J. 1993. PHYLIP (phylogeny inference package), version 3.5 c. Joseph Felsenstein.
- Gehrke PC. 1988. Feeding energetics and angling catches of spangled perch, Leiopotherapon unicolor (Günther 1859), (Percoidei: Teraponidae). Mar Fresh Res. 39:569–577.
- Gehrke PC. 1987. Cardio‐respiratory morphometrics of spangled perch, Leiopotherapon unicolor (Günther, 1859), (Percoidei, Teraponidae). J Fish Biol. 31:617–623.
- Li SF, Yan LP, Li CS, Hu F. 2004. Analysis of fish composition characteristics in the northern part of the east China sea. J Fish China. 28:384–392.
- Lin Q, Dai FQ, Zhang B, Jin XS, Li ZY. 2009. Principal component analysis (pca) for the quality analysis and evaluation of six bait fishes in the yellow sea (in Chinese). Prog Fish Sci. 30:64–68.
- Zhang B, Tang QS, Jin XS, Xue Y. 2005. Food competition for the main fish in the east China sea and the yellow sea (In Chinese). Curr Zool. 51:616–623.