Abstract
The whole chloroplast genome of Amorphophallus konjac is determined in this study. The genome size was 161,647 bp in length, containing a large single copy region of 90k bp and a small single copy region of 20k bp, which were separated by a pair of 25,722 bp inverted repeat regions. The complete chloroplast genome contained 131 genes, including 86 coding-protein (79 PCG species), 8 rRNA (4 rRNA species), and 37 tRNA genes (30 tRNA species). Based on the phylogenetic analysis, the genus Amorphophallus, represented by A. konjac might show a close relationship with Pinellia and Colocasia by the limited data for current. These complete chloroplast genomes of A. konjac laid a good foundation for population genomics, molecular, and ecological studies of Amorphophallus.
Amorphophallus konjac K. Koch, synonym A. rivieri Durieu, has been used in China, Japan, and South East Asia for thousands of years as an important crop plant and traditional medicine (Li and Long Citation1998, Li et al. Citation2010; Chua et al. Citation2010). Currently, there was no complete chloroplast genome sequence of any species from genus Amorphophallus available in the GenBank database. We aimed to assemble and characterize the complete A. konjac plastide genome to provide a better understanding of the evolution and phylogenetic relationship of genus Amorphophallus in Araceae.
One A. konjac individual was collected from Longquan Mountain, Guizhou, China (E107°28′3.68″, N27°46′55.23″). The voucher specimen (WangQ-Anjx-20160611) was deposited in the herbarium of ZMU. Total genomic DNA was isolated using a modified CTAB method (Doyle and Doyle Citation1987) and sequenced by Illumina Hiseq Platformfrom GENEWIZ. A total of 50,159,386 paired-end (150 bp) reads were obtained. After removal of adapter sequences, high-quality clean reads were initially mapped to publish chloroplast genome of Colocasia esculenta (JN105690) using BWA (Li and Durbin Citation2009) and SAMtools (Li et al. Citation2009). Then, we assembled these reads into complete chloroplast genome using Velvet (Zerbino and Birney Citation2008) and filled gaps with GapCloser (http://soap.genomics.org.cn/index.html). Thirdly, we annotated the plastome using Plann (Huang and Cronk Citation2015) and manually corrected the annotation with Generous (Kearse et al. Citation2012) and Sequin (http://www.ncbi.nlm.nih.gov/Sequin/) based on Colocasia esculenta plastome as a reference annotation. The complete plastome sequence together with gene annotations were submitted to GenBank under the accession number MK611803 for A. konjac for the first time.
The complete chloroplast genome of A. konjac was 161,647 bp, consisting of a pair of inverted repeat regions of 25,722 bp each, a large single copy region of 90,006 bp, and a small copy region of 20,197 bp. The overall GC content of the complete plastid genome was 42.3% and it contained 131 genes, including 86 coding-protein (79 PCG species), 8 rRNA (4 rRNA species), and 37 tRNA genes (30 tRNA species). Most of the gene species occurred in a single copy, while 18 gene species occurred in double copies, including all rRNA species (4.5S, 5S, 16S, and 23S rRNA), 7 tRNA species and 7 PCG species (ndhB, rpl2, rps12, rpl23, rps7, ycf2, and ycf68).
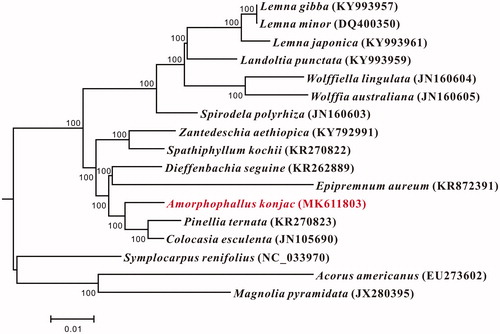
To validate the phylogenetic position of A. konjac, a neighbor-joining (NJ) tree with 500 bootstrap replicates was inferred using MEGA10 (Saitou and Nei Citation1987; Kumar et al. Citation2018) from alignments created by the MAFFT online version (Katoh and Standley Citation2013) containing all published plastid genomes of Araceae species, Magnolia pyramidata, and Acorus americanus were chosen as outgroups (). The phylogenetic analysis indicated that genus Amorphophallus, represented by A. konjac might show a close relationship with Pinellia and Colocasia for current data (). Meanwhile, the phylogenetic relationship of Symplocarpus within Araceae need more species and discussions. The whole chloroplast genome of A. konjac is useful for population genetic studies of Amorphophallus and more species needed to verify the clear phylogenetic relationship.
Figure 1. Neighbor-joining (NJ) tree based on the whole chloroplast genome sequences of 17 taxa including 15 species chloroplast genome sequences of Araceae and Acorus americanus and Magnolia pyramidata were used as the outgroups. Bootstrap support values (%) from 500 replicates are indicated in each node. GenBank accession numbers: Lemna gibba (KY993957), Lemna minor (DQ400350), Lemna japonica (KY993961), Landoltia punctata (KY993959), Wolffiella lingulata (JN160604), Wolffia australiana (JN160605), Spirodela polyrhiza (JN160603), Zantedeschia aethiopica (KY792991), Spathiphyllum kochii (KR270822), Dieffenbachia seguine (KR262889). Epipremnum aureum (KR872391), Amorphophallus konjac (MK611803), Pinellia ternata (KR270823), Colocasia esculenta (JN105690), Symplocarpus renifolius (NC_033970), Acorus americanus (EU273602), and Magnolia pyramidata (JX280395).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chua M, Baldwin TC, Hocking TJ, Chan K. 2010. Traditional uses and potential health benefits of Amorphophallus konjac K. Koch ex N.E.Br. J Ethnopharmacol. 128:268–278.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500023–1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and samtools. Bioinformatics. 25:2078–2079.
- Li H, Long CL. 1998. Taxonomy of Amorphophallus in China. Acta Bot Yunnanica. 20:167–170.
- Li H, Zhu GH, Peter CB, Jin M, Wibert LH, Josef B, Niels J. 2010. Flora of China. Vol. 23. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
