Abstract
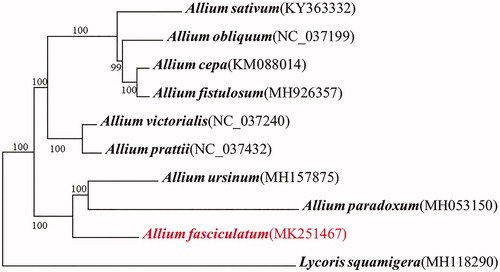
Allium fasciculatum is an endemic alpine species of Allium L., which is confined to the Qinghai-Tibet Plateau, China. In this study, we sequenced the complete plastid genome of A. fasciculatum and firstly reported it. A typical quadripartite structure was detected with 152,931 bp in length and it consists of a pair of inverted repeats with 26,464 bp, separated by small and large single copies with 17,838 bp and 82,167 bp, respectively. The genome contained 132 genes, consisted of 85 coding genes, 38 tRNA genes, and 8 rRNA genes. Phylogenetic analyses based on the whole complete genome sequences showed A. fasciculatum was closely clustered with A. paradoxum and Allium ursinum with high support.
Allium fasciculatum Rendle is a species of Allium L., which is endemic to the Eastern Himalaya–Hengduan Mountains and neighboring areas (Li et al. Citation2010). A. fasciculatum usually grows on dry slopes and sandy places at elevations from 2200 to 5400 m with tuberous, short, thick roots, and umbel globose (Xu and Kamelin Citation2000). We assembled the complete chloroplast genome sequence of A. fasciculatum to inform genomic resources for future studies of phylogenetic analysis.
A. fasciculatum was sampled fresh leaves collected Chayu (Tibet, China; coordinates: 28°38′N, 97°47′E), and the specimen (voucher number: LH20180921-4) has been deposited in Sichuan University Herbarium (SZ). The total genomic DNA was isolated from leaf tissues using a modified cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle Citation1987). The isolated genomic was manufactured to average 350 bp paired-end (PE) library using Illumina Hiseq platform (Illumina, San Diego, CA, USA) and sequenced using Illumina genome platform (HiseqPE150, Illumina, San Diego, CA, USA). The filtered reads were assembled using the program NOVOPlasty (Dierckxsens et al. Citation2017) with its close relative complete chloroplast genome of Allium victoralis as the reference (GenBank accession no. NC_037240). The assembled chloroplast genome was annotated using Geneious 11.0.4 and checked plus manual corrections (Kearse et al. Citation2012). Ten species of Amaryllidaceae were performed on sequence alignments in the complete chloroplast genome sequences. The complete chloroplast genome of 10 species was aligned using MAFFT (Katoh et al. Citation2002). Maximum Likelihood(ML) analyses with 1000 replicates were reconstructed by RAxML 8.2.11 (Stamatakis Citation2006).
The chloroplast genome of A. fasciculatum was 152,931 bp in length (GenBank accession no. MK251467), containing a large single copy region (LSC) of 82,167 bp, a small single copy region (SSC) of 17,838 bp, and a pair of inverted repeat (IR) regions of 26,464 bp. Genome annotation predicted 132 genes, including 85protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The overall GC-content of the whole plastome was 37.1%, with the corresponding values in the LSC, SSC, and IR regions were34.9, 30.1, and 42.7%, respectively. In the IR regions, it was apparently higher than in the other regions, possibly because of the high GC-content of the rRNA (55.8%).
Phylogenetic analyses based on the whole complete genome sequences showed A. fasciculatum was closely clustered with A. paradoxum and Allium ursinum with high support (), which was consistent with the previous study (Li et al. Citation2010). All the phylogenetic analysis is substantially increasing our understanding of the evolutionary relationship among species in Allium.
Acknowledgments
We acknowledge Yan Yu and Jun-Pei Chen for the help in technical support and materials collection.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bul. 19:11–15.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Li QQ, Zhou SD, He XJ, Yu Y, Zhang YC, Wei XQ. 2010. Phylogeny and biogeography of Allium (Amaryllidaceae: Allieae) based on nuclear ribosomal internal transcribed spacer and chloroplast rps16 sequences, focusing on the inclusion of species endemic to China. Ann Bot. 106:709–733.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Xu JM, Kamelin RV. 2000. Allium L. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 24. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 165–202.