Abstract
Nilaparvata lugens, called as brown planthoppers (BPH), is one of the important pests on rice. To identify the origin of Korean N. lugens, we completed the mitochondrial genome of N. lugens captured in Guangdong province, China. The circular mitogenome of N. lugens is 17,606 bp including 13 protein-coding genes, 2 rRNAs, 22 tRNAs, and a single large non-coding region of 2,424 bp. The base composition was AT-biased (89.5%). Ninety single nucleotide polymorphisms and ten insertions and deletions are identified by comparing with Korean N. lugens. Phylogenetic trees present that Guangdong may not be an origin of Korean BPH based on the distance of two mitochondrial genomes.
Nilaparvata lugens (Stål, 1854) is one of the important rice pests, called brown planthoppers (BPH; Sogawa and Cheng Citation1979). It is widely distributed in Asia, Northern Australia, and South Pacific archipelago, where rice has been cultivated widely (Hinckley Citation1963; Abraham and Nair Citation1975). In 2013, the first BPH mitochondrial genome was analyzed together with Laodelphax striatellus, displaying short Atp8 and rearrangement of tRNAs (Zhang et al. Citation2013). Additional mitochondrial genomes of biotype 1, 2, and 3, and Nilaparvata muiri were sequenced, showing Nad2 is the most variables among N. lugens mitochondrial genomes (Lv et al. Citation2015). Consequently, mitochondrial genomes of two more biotypes, Y and L, were also sequenced. Moreover, N. lugens captured in Korea was analyzed to find its origin due to periodic migration from China to Korea (Kim et al. Citation1985; Saxena and Barrion Citation1985), presenting its mitochondrial genome that is positioned outside of clade of N. lugens (Park et al., Citationunder review).
To understand intra-species variations of mitochondrial genome of N. lugens more, total DNA of N. lugens captured at Guangdong province (23°03′18.74′N, 114°35′11.58′E), China, was obtained (Specimen was deposited at InfoBoss Herbarium (IN), Korea; Choi NJ, INH-00015) was extracted using DNA extraction method manually (Zymo Research, DNA extraction kit, USA). Sequencing was conducted by HiSeqX (Macrogen Inc., Korea) and de novo assembly and confirmation were done by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate mitochondrial genome based on N. lugens mitochondrial genome (MK590088; Park et al., under revision) together with ARWEN for tRNAs (Laslett and Canbäck Citation2008).
Nilaparvata lugens mitochondrial genome (Genbank accession is MK606371) is 17,606 bp, which is shorter than that of N. lugens captured in Korea (MK590088; Park et al., Citationunder review) by 4 bp. It contains 13 protein-coding genes, 2 rRNAs, and 22 tRNAs. Its nucleotide composition is AT-biased (A + T ratio is 77.1%). The control region, presumably single largest non-coding AT-rich region, is 2,424 bp (AT ratio is 80.7%). Ninety single nucleotide polymorphisms (SNPs) and 10 insertions and deletions are identified by comparing with that of Korean BPH, which is a similar level of variation between Korean BPH and its first mitochondrial genome (Park et al., Citationunder review) and is lower than those of Chilo suppressalis (Park et al. Citation2019). Seventeen synonymous SNPs in eight genes and two non-synonymous SNPs and one deletion in ND5, CTYB, and ND1, respectively, are identified.
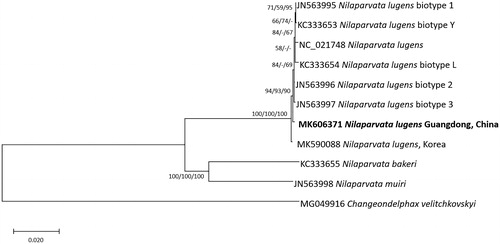
We inferred the phylogenetic relationship from 11 Delphacidae complete mitochondrial genomes aligned by MAFFT 7.388 (Katoh and Standley Citation2013). Neighbor joining, minimum evolution, and maximum likelihood bootstrapped phylogenetic trees were constructed using MEGA X (Kumar et al. Citation2018). Phylogenetic trees present that five mitochondrial genomes with identified biotypes are not clustered in one clade with those of Guangdong and Korean BPHs, presenting distance between the two mitochondrial genomes and the rest six (). In addition, Guangdong BPH is also distinct from Korean PBH, suggesting that the origin of Korean BHP may not be Guangdong area, China ().
Figure 1. Neighbor joining (bootstrap repeat: 10,000), minimum evolution (bootstrap repeat: 10,000), and maximum likelihood (bootstrap repeat: 1,000) phylogenetic trees of fifteen Delphacidae and one Fulgoridae complete mitochondrial genomes: Eight Nilaparvata lugens (MK606371 in this study, MK590088, JX880069, JN563995, KC333653, JN563996, JN563997, and KC333654), Nilaparvata muiri (JN563998), Nilaparvata bakeri (KC333655), and Changeondelphax velitchkovskyi (MG049916) as an outgroup species. Phylogenetic tree was shown based on neighbor joining tree. The numbers above branches indicate bootstrap support values of neighbor joining, minimum evolution, and maximum likelihood phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abraham C, Nair M. 1975. The brown planthopper outbreaks in Kerala, India. Rice Entomol Newsl. 2:36.
- Hinckley AD. 1963. Ecology and control of rice planthoppers in Fiji. Bull Entomol Res. 54:467–481.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim Y, Lee J, Park H, Kim M. 1985. Plant damages and yields of the different rice cultivars to brown planthopper (Nilaparvata lugens S.) in fields. Korean J Appl Entomol. 24:79–83.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Lv L, Peng X, Jing S, Liu B, Zhu L, He G. 2015. Intraspecific and interspecific variations in the mitochondrial genomes of Nilaparvata (Hemiptera: Delphacidae). J Econ Entomol. 108:2021–2029.
- Park J, Kwon W, Park J, Kim HJ, Lee B-C, Kim Y, Choi NJ. under review. The complete mitochondrial genome of Nilaparvata lugens (Stål, 1854) captured in Korea (Hemiptera: Delphacidae). DOI:10.1080/23802359.2019.1606680
- Park J, Xi H, Kwon W, Park C-G, Lee W. 2019. The complete mitochondrial genome sequence of Korean Chilo suppressalis (Walker, 1863)(Lepidoptera: Crambidae). Mitochondr DNA B. 4:850–851.
- Saxena R, Barrion A. 1985. Biotypes of the brown planthopper Nilaparvata lugens (Stål) and strategies in deployment of host plant resistance. Int J Trop Insect Sci. 6:271–289.
- Sogawa K, Cheng C. 1979. Economic thresholds, nature of damage, and losses caused by the brown planthopper. Brown planthopper: Threat to rice production in Asia. Manila: International Rice Res Institute; p. 125–142.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genom Res. 18:821–829.
- Zhang K-J, Zhu W-C, Rong X, Zhang Y-K, Ding X-L, Liu J, Chen D-S, Du Y, Hong X-Y. 2013. The complete mitochondrial genomes of two rice planthoppers, Nilaparvata lugens and Laodelphax striatellus: conserved genome rearrangement in Delphacidae and discovery of new characteristics of atp8 and tRNA genes. BMC Genomics. 14:417.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC bioinformatics. 12(suppl14):S2.
