Abstract
In this study, we sequenced the complete chloroplast genome of Dracaena cochinchinensis and used the data to assess its genomic resources. The complete chloroplast genome revealed a circular genome of 155,182 bp size with 37.5% GC content. The chloroplast genome is a typical quadripartite chloroplast structure with inverted repeats (26,507 bp) separated by small single copy (18,466 bp) and large single copy (83,702 bp) regions. De novo assembly and annotation showed the presence of 112 unique genes with 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. We identified a total of 68 simple sequence repeat (SSR) markers in the chloroplast genome. A maximum-likelihood phylogenomic analysis showed that the position of D. cochinchinensi was on the base of Nolinoideae. Overall, the results of this study will contribute to better support of the evolution, molecular biology and conservation of D. cochinchinensi.
The plant Dracaena cochinchinensis Lour. has been used as a traditional medicine since ancient times in the form of Dragon’s blood, a deep red resin (Khieu et al. Citation2015). Over the past several decades, due to its high market demand, overexploitation, habitat destruction, and ecosystem deterioration caused a rapid decline in the natural resource of D. cochinchinensis. D. cochinchinensi has been listed as a class II conserved medicinal plant in China. Here, we sequenced and analyzed the chloroplast genome of D. cochinchinensi based on the Illumina pair-end sequencing data. Our aim was to retrieve chloroplast genome structure and valuable genomic resources for this species.
Fresh leaves of D. cochinchinensi were collected from Beijing Botanical Garden, Chinese Academy of Sciences (N39°59′15.75″, E116°12′34.58). The specimen is stored at PE. Total genomic DNA was extracted and purified following the method of (Li et al. Citation2013). The sequencing library was constructed and quantified following the methods introduced by (Dong et al. Citation2017). Paired-end sequencing (2 × 150 bp) was conducted on an Illumina HiSeq 4000 platform; reads were qualitatively assessed and assembled with SPAdes 3.6.1 (Bankevich et al. Citation2012). The depth of average genome coverage exceeded 1621X. Gene annotation of D. cochinchinensi was performed using DOGMA annotation (Wyman et al. Citation2004) and manually corrected for codons and gene boundaries using BLAST searches. The annotated genomic sequence had been submitted to GenBank with the accession number MF943127.
The genome of D. cochinchinensi is a circular molecule of 155,182 base pairs (bp), with a pair of Inverted Repeats (IR) of 26,507 bp each, separated by two single copies: a large (LSC, 83,702 bp) and a small single copy (SSC, 18,466 bp). D. cochinchinensi chloroplast genome has a 37.5% GC content and encodes 112 unique genes, 78 of which code for proteins, 30 are tRNA genes and four are rRNA genes, which is consistent with those from other species in Agavaceae.
In this study, a total of 68 perfect SSR were detected in D. cochinchinensi chloroplast genome using a Perl script MISA, including 42 mononucleotides, 14 dinucleotides, two trinucleotides, nine tetranucleotides, and one hexanucleotide. Most SSRs were located in the intergenic region with the A or the T motif.
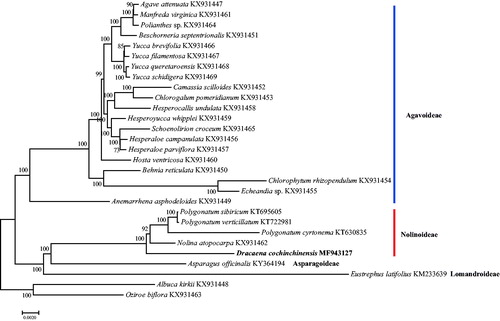
To obtain insight into the position of D. cochinchinensi within the Asparagaceae, we generated datasets of 81 protein-coding genes of 29 species, using Albuca kirkii and Oziroe biflora as outgroups (). Phylogenetic analyses were performed using maximum likelihood (ML) in RAxML8.0 with 1000 bootstrap replicates (Stamatakis Citation2014). ML analysis revealed that most of the nodes had 100% bootstrap value. The position of D. cochinchinensi is on the base of Nolinoideae. The complete chloroplast genome provides interesting insights and valuable genomic resources of D. cochinchinensi that can be used to reconstruct its phylogeny, evaluate genetic variations, and develop conservation strategy.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Dong W, Xu C, Li W, et al. 2017. Phylogenetic resolution in Juglans based on complete chloroplast genomes and nuclear DNA sequences. Front Plant Sci. 8:1148.
- Khieu T-N, Liu M-J, Nimaichand S, Quach N-T, Chu-Ky S, Phi Q-T, Vu T-T, Nguyen T-D, Xiong Z, Prabhu DM, et al. 2015. Characterization and evaluation of antimicrobial and cytotoxic effects of Streptomyces sp. HUST012 isolated from medicinal plant Dracaena cochinchinensis Lour. Front Microbiol. 6:574.
- Li J, Wang S, Jing Y, Wang L, Zhou S. 2013. A modified CTAB protocol for plant DNA extraction. Chinese Bull Bot. 48:72–78.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.