Abstract
The mitochondrial genome of Issoria eugenia is described in this study. The molecule is 15,206 bp in length, containing 37 typical insect mitochondrial genes and one AT-rich region. All protein-coding genes (PCGs) started with ATN, except for COI gene with CGA, nine PCGs harbour the typical stop codon TAA, whereas four PCGs terminate with TA or T. The rrnL and rrnS genes are 1316 bp and 736 bp in length, respectively. The AT-rich region contains several non-repetitive sequences and several structural features characteristic of lepidopterans. The mitogenome data would be useful for further understanding the taxonomy and phylogeny of Heliconiinae.
The subfamily Heliconiinae (Lepidoptera: Nymphalidae) is composed of a diverse group of species distributed widely in the New and Old World tropics (Brower Citation2000; Penz and Peggie Citation2003). However, their taxonomy and phylogeny are still standing as a controversial issue (Wahlberg et al. Citation2009; Shi et al. Citation2015). In recent decades, the insect mitochondrial genomes (mitogenomes) have been widely used as an informative molecular marker for phylogenetic and population genetic studies at various hierarchical levels, due to their unique features (Boore Citation1999; Salvato et al. Citation2008; Wu et al. Citation2013).
Here, we sequenced the complete mitochondrial genome of Issoria eugenia, a commonly found member of subfamily Heliconiinae in Nymphalidae. Adult individuals of Issoria eugenia were collected at Qilian Mountain, Gansu province, China (coordinates: E99.476, N38.612) on July 2016. After morphological identification, fresh samples were preserved in 100% ethanol and kept in the Laboratory of Molecular Phylogenetics and Evolution (Anhui Normal University, China) at −20 °C until use. Total genomic DNA was extracted from the thorax muscle of a single individual using the Sangon Animal genome DNA Extraction Kit (Shanghai, China). The resultant reads were assembled and annotated using the BioEdit 7.0 (Hall, Citation1999) and MEGA 7.0 softwares (Kumar et al. Citation2016).
The whole mitogenome is a circular molecule of 15,206 bp in size (GenBank accession No. MK598743), and contains 13 protein-coding genes (PCGs), 22 transfer tRNA genes and 2 ribosomal rRNA genes, along with an AT-rich control region. Its gene arrangement and orientation are identical to those of other known nymphalid mitogenomes (Wu et al. Citation2013; Mccullagh and Marcus Citation2015; Shi et al. Citation2015). The A + T content of the mitogenome is 80.3%, which is generally in accordance with other nymphalid mitogenomes. All PCGs are initiated by typical ATN codons, except for COI gene, which starts with the unusual CGA as observed in most of the other sequenced nymphalids (Gan et al. Citation2014; Shi et al. Citation2015). Nine PCGs have a complete stop codon TAA, whereas four PCGs (COI, COII, ND1 and ND4) stop with incomplete TA or T. All tRNAs harbour the typical predicted secondary cloverleaf structures except for the tRNA−Ser(AGN), as seen in other determined butterflies (Hao et al. Citation2013; Shi et al. Citation2015). The rrnL and rrnS genes are 1316 bp and 736 bp in length, respectively. The AT-rich region is 402 bp long with several structures characteristic of lepidopterans.
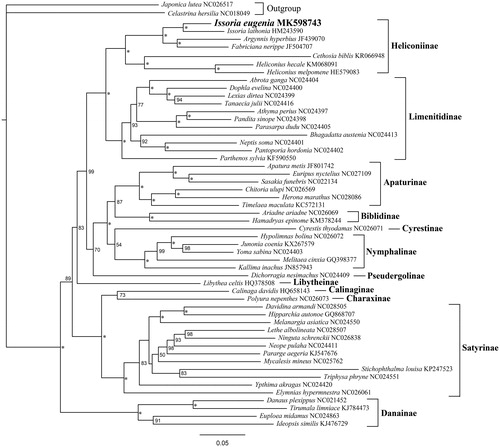
We performed a maximum-likelihood (ML) phylogenetic analysis with IQ-TREE (version 1.6.1) based on nucleotides sequences of 13 PCGs to understand the phylogenetic relationship of Issoria eugenia with other 51 nymphalids available from GenBank (see for details), using 2 Lycaenidae species as outgroups. The resultant ML tree exhibited that Issoria eugenia constituted a monophyletic group with other Nymphalinae species, and Issoria eugenia is shown to be clustered within the Heliconiinae group with strong support value. In addition, the Heliconiinae of this study includes five genera, namely the Issoria, Argynnis, Fabriciana, Cethosia, and Heliconius, with their relationship of ((Cethosia + Heliconius) + ((Argynnis + Fabriciana) + Issoria)).
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this paper.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Brower AV. 2000. Phylogenetic relationships among the Nymphalidae (Lepidoptera) inferred from partial sequences of the wingless gene. Proc Royal Soc London Series B: Biol Sci. 267:1201–1211.
- Gan S, Chen Y, Zuo N, Xia C, Hao J. 2014. The complete mitochondrial genome of Tirumala limniace (Lepidoptera: Nymphalidae: Danainae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1096–1098.
- Hall, TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Hao J, Sun M, Shi Q, Sun X, Shao L, Yang Q. 2013. Complete mitogenomes of Euploea mulciber (Nymphalidae: Danainae) and Libythea celtis (Nymphalidae: Libytheinae) and their phylogenetic implications. ISRN Genomics. 2013:1.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Mccullagh BS, Marcus JM. 2015. The complete mitochondrial genome of the lemon pansy Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae). J Asia-Pacific Entomol. 18:749–755.
- Penz CM, Peggie D. 2003. Phylogenetic relationships among Heliconiinae genera based on morphology (Lepidoptera: Nymphalidae). System Entomol. 28:451–479.
- Salvato P, Simonato M, Battisti A, Negrisolo E. 2008. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genomics. 9:331.
- Shi QH, Sun XY, Wang YL, Hao JS, Yang Q. 2015. Morphological characters are compatible with mitogenomic data in resolving the phylogeny of nymphalid butterflies (Lepidoptera: Papilionoidea: Nymphalidae). PLoS One. 10:e0124349.
- Wahlberg N, Leneveu J, Kodandaramaiah U, Peña C, Nylin S, Freitas AV, Brower AV. 2009. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc Royal Soc B: Biol Sci. 276:4295–4302.
- Wu Q-L, Gong Y-J, Gu Y, Wei S-J. 2013. The complete mitochondrial genome of the beet armyworm Spodoptera exigua (Hübner) (Lepodiptera: Noctuidae). Mitochondrial DNA. 24:31–33.