Abstract
The complete chloroplast genome sequence of Aquilegia rockii was assembled and characterized as a resource for future genetic studies. With a total length of 162,123 bp, the chloroplast genome comprised of a large single-copy (LSC) region of 91,337 bp, a small single-copy (SSC) region of 17,376 bp, and 2 inverted repeat (IR) regions of 26,705 bp. A total of 117 unique genes, including 81 protein-coding genes, 4 ribosomal RNA genes, and 32 tRNA genes. The maximum likelihood phylogenetic analysis based on 18 cp genomes indicated that A. rockii is sister to the clade formed by Semiaquilegia adoxoides and Urophysa rockii.
The genus Aquilegia known for the spurred petals of their flowers consists of ∼75 species with a similar number of taxa occurring in North America (22 spp), Asia (23 spp), and Europe (21 spp) (Nold Citation2003). Their petals show an enormous range of spur length diversity ranging from 1 to 15 cm spurs of Aquilegia longissima (Puzey et al. Citation2011). Selection from pollinator shifts is suggested to have driven these changes in nectar spur length (Whittall and Hodges Citation2007). Consequently, the genetic and genomic information of Aquilegia is needed to evaluate how the pollinator-mediated selection drives speciation. In this study, we made the first report of a complete plastome for A. rockii, a columbine in western China (GenBank accession number: MK573514).
The leaves of A. rockii were collected from Hong Shan, Shangri-La, Yunnan, China (28°15'N, 100°5'E) and the specimen was deposited at the herbarium of Kunming Institute of Botany, Chinese Academy of Science. Total genomic DNA was extracted from the silica-dried leaves with a modified CTAB method (Doyle and Doyle Citation1987). The genomic paired-end (PE150) sequencing was performed on an Illumina Hiseq 2000 instrument (Illumina, San Diego, CA, USA). The cp genome was assembled using the program NOVOPlasty (Dierckxsens et al. Citation2017) with the partial chloroplast sequence of A. rockii (KC288600.1) as seed. Annotation was performed using PGA (Qu Citation2019).
The complete chloroplast genome of A. rockii is 162,123 bp in length and contains two inverted repeat (IRa and IRb) regions of 26,705 bp, which was separated by a large single-copy (LSC) region of 91,337 bp and a small single-copy (SSC) region of 17,376 bp (). The overall GC content was 39.0%, while higher in IR (43.0%) than in LSC (37.7%) and SSC (33.4%). The new sequence possesses a total of 117 unique genes, including 81 protein-coding genes, 4 ribosomal RNA genes, and 32 tRNA genes. In these genes, five tRNA genes (i.e. trnA-UGC, trnI-GAU, trnK, trnL-UAA, and trnV-UAC) and nine protein-coding genes (atpF, ndhA, ndhB, petB, petD, rps12, rps16, rpl2, and rpoC1) contained one intron, and the ycf3 gene has two introns. Five protein-coding genes, seven tRNAs and all four rRNAs were completely duplicated within IRs.
Figure 1. ML phylogenetic tree of 16 species within Ranunculaceae based on 15 chloroplast genome sequences in GenBank, plus the chloroplast sequence of Aquilegia rockii. The tree is rooted with the Berberidaceae (Ranzania japonica and Berberis amurensis). Bootstraps (10,000 replicates) are shown at the nodes.
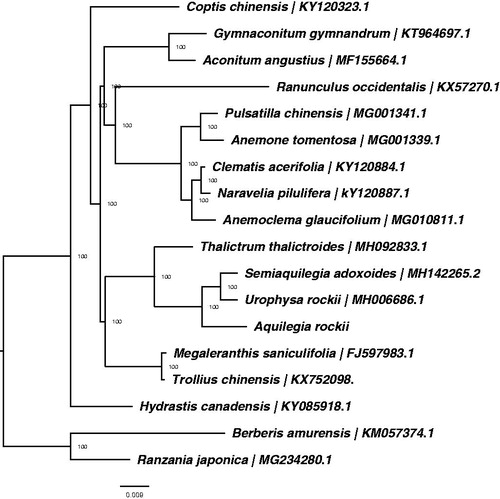
To understand the phylogenetic position of A. rockii within the family Ranunculaceae, we downloaded the complete chloroplast genomes of 17 species from the NCBI GenBank database, including 15 species in Ranunculaceae and two species in Berberidaceae. The sequences were aligned using MAFFT v7.307 (Katoh and Standley Citation2013), and RAxML (Stamatakis Citation2014) was used to construct a maximum likelihood tree with Ranzania japonica and Berberis amurensis as outgroups. All nodes in the complete plastome tree were strongly supported. The phylogenetic tree showed that A. rockii was closely related to the clade including Semiaquilegia adoxoides and Urophysa rockii (). This published A. rockii chloroplast genome will provide useful information for phylogenetic and evolutionary studies in Ranunculaceae and Aquilegia.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Nold R. 2003. Columbines. Aquilegia, paraquilegia and semiaquilegia. Cambridge (UK): Timber Press.
- Puzey JR, Gerbode SJ, Hodges SA, Kramer EM, Mahadevan L. 2011. Evolution of spur-length diversity in Aquilegia petals is achieved solely through cell-shape anisotropy. Proc R Soc Lond B. 279:1640–1645.
- Qu X-J.2019. Plastid Genome Annotator. GitHub. [Accessed 2019 Jan 15]. https://github.com/quxiaojian/PGA.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Whittall JB, Hodges S. 2007. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature. 447:706–709.
