Abstract
IIn this study, the complete mitochondrial genome of the Sternaspis scutata wassequenced. The mitochondrial genome is 15 300 bp in length, containing 13 protein-coding sequences, 2 rRNAs, 21 tRNAs and 1 non-coding region. All these genes are in the heavy strand. Most protein-coding genes begin with an ATG codon, while NAD4L utilize ATT. The common termination codon is TAA, except for atp6 terminated with TAG, and cox2, atp8, nad2, nad6 terminated by incomplete codon T. Seen from the phylogenetic tree, Sternaspis scutata, Pista cristata and Terebellides stroemii from the same Order (Terebellida) clustered into one branch.
Sternaspis scutata belongs to the family Sternaspidae and characterized by its peanut-like body shape and a remarkable shield that lies ventro-posteriorly. Sternaspid polychaetes are very common and frequently dominant in the sea areas of China (Wang et al. Citation2006). Some authors suggest that only one species should be recognized (Yang and Sun Citation1988), whereas others regard a few species (Wu et al. Citation2015; Wu and Xu Citation2017). In this paper, we present the complete mitochondrial genome of S. scutata by using the next-generation sequencing (NGS) techniques strategy (Wang et al. Citation2015), which can help to unveil taxonomic problems and better understand the phylogenetic status of this species in Phylum Annelida. The S. scutata specimen used in this study was collected from Jiaozhou Bay (36.11°N, 120.19°E), during October 2018. All examined specimens were preserved at the Marine Ecology Research Center of the First Institute Oceanography, Ministry of Natural Resources in Qingdao.
The complete mitochondrial genome of S. scutata (GenBank accession no. MK548645) is 15,300 bp in length, containing 13 protein-coding sequences (CDS), 2 ribosomal RNA genes (12SrRNA and 16S rRNA), 21 transfer RNA genes (tRNA) and 1 non-coding control region (D-loop). The CDS sequences correspond to the genes cox1, cox2, cox3, atp6, atp8, nad6, cob, nad1, nad5, nad4l, nad4, nad3, and nad2. All these genes are in the heavy strand. The 2 rRNA and 21 tRNA genes are distributed in the heavy strand, too. The overall nucleotides base composition of the heavy strand is 28.73% for A, 25.20% for C, 11.99% for G and 34.08% for T, with a slight A + T-rich feature (62.81%). Most protein-coding genes begin with ATG, except for NAD4L, which started with ATT. The cox1, cox3, cob, nad1, nad3, nad4, nad4l, and nad5 are terminated with TAA, atp6 is terminated with TAG, while cox2, atp8, nad2, nad6 are stopped by incomplete codon T. The longest one is nd5 gene (1710 bp) among the protein-coding genes, whereas the shortest is atp8 gene (160 bp). The non-coding control region (D-loop) is 503 bp in length and is located between tRNA-Cys (GCA) and tRNA –Thr (TGT). The two ribosomal RNA genes, 12S rRNA (833 bp) and 16S rRNA (1289 bp), are located between the tRNA-Met (CAT) and tRNA-Leu (TAG) genes and are further separated by the tRNA-Val (TAC) gene.
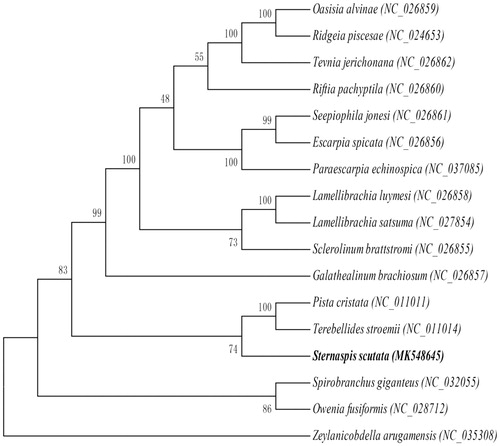
The maximum likelihood tree (by RAxML 8.1.5, Heidelberg, Germany) (Alexandros Citation2006) based on mitochondrial genome nucleotide sequences of the S. scutata and the other 16 species of Polychaeta were constructed under the mutation model of GTR + I + G (by jModelTest 2.1.7, Vigo, Spain) (David Citation2008). This is the first record of the complete mitochondrial genome of genus Sternaspis. But in the Order Terebellida, there have been other two recorded complete mitochondrial genome, i.e., Pista cristata and Terebellides stroemii. According to the phylogenetic analysis, S. scutata has a more close relationship with P. cristata and T. stroemii than other species ().
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Figure 1. Maximum likelihood tree based on mitochondrial genome nucleotide sequences of the Sternaspis scutata (MK548645) and the other 16 species of Polychaeta under the mutation model of GTR + I + G. Genbank accession numbers of the sequences were used for the tree as follows: Pista cristata (NC_011011.1), Terebellides stroemii (NC_011014.1), Paraescarpia echinospica (NC_037085.1), Spirobranchus giganteus (NC_032055.1), Owenia fusiformis (NC_028712.1), Lamellibrachia satsuma (NC_027854.1), Ridgeia piscesae (NC_024653.1), Tevnia jerichonana (NC_026862.1), Galathealinum brachiosum (NC_026857.1), Seepiophila jonesi (NC_026861.1), Sclerolinum brattstromi (NC_026855.1), Lamellibrachia luymesi (NC_026858.1), Riftia pachyptila (NC_026860.1), Oasisia alvinae (NC_026859.1), Escarpia spicata (NC_026856.1), Zeylanicobdella arugamensis (NC_035308.1).
Additional information
Funding
References
- Alexandros S. 2006. RAxML-VI-HPC: maximum likelihood-basedphylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- David P. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256.
- Wang JB, Li XZ, Wang HF. 2006. Ecological characteristics of dominant polychaete species from the Jiaozhou Bay. Acta Zoologica Sinica. 52:63–69.
- Wang Q, Wang J, Luo J, Chen GH. 2015. The complete mitochondrial genome of the Synanceia verrucosa (Scorpaeniformes: Synanceiidae). Mitochondrial DNA. 26:1–2.
- Wu XW, Sergio IS, Xu KD. 2015. Two new species of Sternaspis Otto, 1821 (Polychaeta: Sternaspidae) from China seas. Zootaxa. 4052:373–382.
- Wu XW, Xu KD. 2017. Diversity of Sternaspidae (Annelida: Terebellida) in the South China Sea, with descriptions of four new species. Zootaxa. 4244:403–415.
- Yang DJ, Sun RP. 1988. Polychaetous Annelids commonly seen from the Chinese Waters. Beijing, China: China Agriculture Press; p. 352.
