Abstract
Zanthoxylum nitidum (Roxb.) DC. and Xanthoceras sorbifolium Bunge are important economic species in Sapindales. Here, we report two complete plastid genome sequences of Z. nitidum and X. sorbifolium. The plastid genomes were found to be 157,307 bp and 160,002 bp in length and GC contents were 38.5 and 36.7%, respectively. In addition, we also used the genome skimming method in Nitraria sibirica and Kirkia wilmsii to get a fully resolved phylogenetic framework for Sapindales.
Introduction
Zanthoxylum nitidum (Roxb.) DC. is a classic traditional Chinese medicinal (TCM) plant distributed from China and India to northern Australia (Deyun et al. Citation1996; Commission Citation2015), which is also called ‘liangmianzhen’ in China. Xanthoceras sorbifolium Bunge is a single species of the Xanthoceras genus, naturally distributed in China (Zhou and Liu Citation2012). This species has high oil content in its seeds, which can be widely used as industrial materials (Liu et al. Citation2013).
The fresh leaves of Z. nitidum and X. sorbifolium were collected from Longzhou, Guangxi (N 22°28′44″, E 107°17′70″), and Fuxin, Liaoning (N 42°16′42″, E 121°17′24″). Both voucher specimens were deposited in the Herbarium of Beijing Forestry University (BJFC) (collection numbers He 2019003 and Xiang 20170706). Genomic DNAs were extracted using the CTAB method (Doyle and Doyle Citation1987), and 2 × 150 bp pair end sequencing was performed on an Illumina HiSeq 4000 platform at Majorbio (http://www.majorbio.com/, China). In addition, we obtained species of N. sibirica (SRR5585869) and K. wilmsii (ERR2040467) RNA-seq raw data from the NCBI SRA database for processing a genome skimming analysis.
For the raw reads of Z. nitidum and X. sorbifolium, de novo assembly was performed using Geneious prime (Kearse et al. Citation2012). The raw reads were mapped to contigs using the Fine Tuning Map program in Geneious prime (iterating up to 100 times) to fill gaps. Complete plastid genomes were annotated using the Plann 1.1.2 (Huang and Cronk Citation2015). The plastid genome sequences were uploaded to GenBank using Bankit. For genome skimming analysis, we mapped N. sibirica and K. wilmsii clean reads to Arabidopsis thaliana plastid genome using the Geneious prime (Kearse et al. Citation2012). All sequences aligned by using MAFFT v7.309. Maximum likelihood (ML) analysis was carried out in RAxML v8.2.12 (Stamatakis Citation2014) with bootstrap 1000 replicates under the GTR + G model and Bayesian inference (BI) was performed with MrBayes v3.2.3 (Ronquist and Huelsenbeck Citation2003) under the GTR + G+I model.
The plastome sequence of Z. nitidum (GenBank accession number MK613864) was 157,307 bp in length containing a large single copy (LSC) region of 84,379 bp, a small single copy (SSC) region of 17,636 bp, and two inverted repeats (IR) of 27,646 bp. The genome contained 113 unique functional genes, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. For X. sorbifolium (GenBank accession number MK613863), the plastome sequence was 160,002 bp in length. The LSC, SSC, and IR regions were 86,880 bp, 18,634 bp, and 27,244 bp, respectively. The genome contained 112 unique functional genes, including 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes.
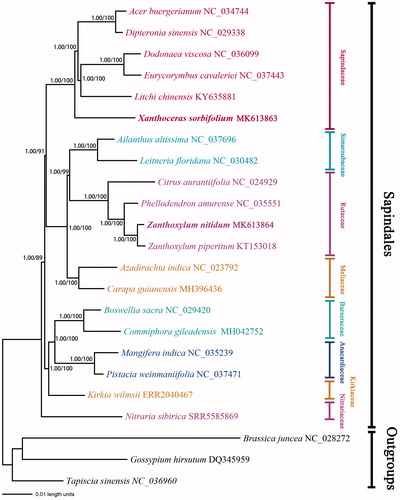
We obtained an almost fully resolved phylogenetic framework for Sapindales (). Support for the sister relationship of Nitrariaceae to the rest of the families of Sapindales. The remaining families are divided into two clades. One of the clades contains three families: (Kirkiaceae, (Burseraceae, Anacardiaceae)), the other clade contains four families (Sapindaceae, (Meliaceae, (Simaroubaceae, Rutaceae))), and Sapindaceae is a sister to the three families.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Commission CP. 2015. Chinese Pharmacopoeia. 2015 ed. Beijing: China Medical Science Press.
- Deyun K, Gray AI, Hartley TG, Waterman PG. 1996. Alkaloids from an Australian accession of Zanthoxylum nitidum (Rutaceae). Biochem Syst Ecol. 24:87–88.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk Q. 2015. Plann: A command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Liu YL, Huang ZD, Ao Y, Li W, Zhang ZX. 2013. Transcriptome analysis of yellow horn (Xanthoceras sorbifolia Bunge): a potential oil-rich seed tree for biodiesel in China. PLoS ONE. 8:e74441.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Zhou QY, Liu GS. 2012. The embryology of Xanthoceras and its phylogenetic implications. Plant Syst Evol. 298:457–468.