Abstract
Siraitia grosvenorii belongs to Cucurbitaceae. It is widely used in beverage raw materials and traditional Chinese medicine. In this study, Illumina sequencing method was used to establish its complete chloroplast (cp) genome. The complete cp genome is 157,132 bp in length, with a large single copy region (LSC) of 92,442 bp and a small single copy region (SSC) of 21,232 bp, which were separated by a pair of inverted repeat (IR) regions of 21,729 bp. The complete cp genome consists of 85 coding sequences (CDS), 39 tRNA, and eight rRNA genes. Phylogenetic analysis showed that S. grosvenorii was clustered into Cucurbitaceae.
Siraitia grosvenorii [Swingle] A. M. Lu et Z. Y. Zhang is a perennial herb of Cucurbitaceae. It is native to in southern China (Pawar et al. Citation2013). Its fruit is called “Luohanguo” and has long been widely used as food and beverage raw materials and traditional Chinese medicine. Its fruit contains non-caloric sweetener mogroside, a group of terpene glycosides (Matsumoto et al. Citation1990, 259) which are about 300 times sweeter than sucrose (Nie Citation1994) and can be used as sucrose substitutes for diabetics and obese patients (Yan et al. Citation2008). Siraitia grosvenorii has been cultivated in Guilin, Guangxi, China for more than 200 years (Lu and Zhang Citation1984).
In this study, Young leaves of S. grosvenorii were collected from Yongfu (Guilin, Guangxi, China; 109°48′E,25°19′N). Plant voucher specimens were kept in herbarium of Guangxi Institute of Botany (IBK; specimen code Xiebingbin201801). The total genomic DNA was extracted from dried leaves using a modified CTAB method (Doyle and Doyle Citation1987) and sequenced based on the Illumina pair-end technology. The filtered reads were assembled using the program Mitobimv1.8 (Hahn et al. Citation2013, 271) and SOAP DeNovo V2.04 (Luo et al. Citation2012, 269) with complete chloroplast genome of Momordica charantia (MG022622.1), Cucumis sativus (DQ865976.1), and Cucumis melo subsp. melo (JF412791) were used as references. Annotated assembled chloroplast genomes using Plann (Huang and Cronk Citation2015) and corrected annotations using Geneious (Kearse et al. Citation2012). Dual Organellar Genome Annotator (DOGMA) software was employed to perform annotation of the protein-coding genes, tRNAs and rRNAs (Wyman et al. Citation2004, 272). The physical map of the chloroplast genome was drawn with OGDRAW (Lohse et al. Citation2013).
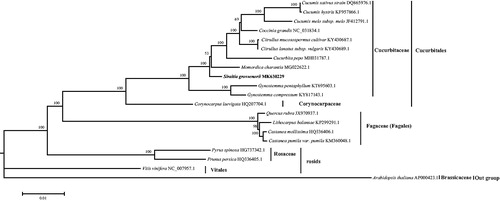
The complete chloroplast genome sequence of S. grosvenorii (GenBank accession MK630229) was 157,132 bp in length. The round genome of S. grosvenorii shows a tetragonal structure, including 105 genes and 45 intergenic spacer regions (IGS). Among 105 genes, 85 are coding sequences (CDS), 39 are tRNAs, and eight are rRNAs genes. Specifically, it has a pair of reverse repeats, Ira, and IRb, both of which are 21,729 bp in size. IRa and IRb repeat sequences are separated by a large single copy (LSC) region and a small single copy (SSC) region, the sizes of which are 92,442 and 21,232 bp, respectively. GC content of the complete cp genome was 36.861%. A total of 20 cp genome sequences were aligned using MAFFT v.7 (Kearse et al. Citation2012) to infer the phylogenetic relationships among the main representative species of Cucurbitales and related orders, and RAxML (Stamatakis Citation2014) was used to construct a maximum likelihood tree with Arabidopsis thaliana as outgroup. As shown in , S. grosvenorii clustered within Cucurbitaceae. The complete chloroplast genome sequence of S. grosvenorii reported here will provide necessary genome information for the protection of germplasm resources of this Cucurbitaceae family member.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle J, Doyle J. 1987. Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Huang DI, Cronk QC. 2015. Plann: a command line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Lu AM, Zhang ZY. 1984. The genus Siraitia Merr. in China. Guihaia. 1:005.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Matsumoto K, Kasai R, Ohtani K, Tanaka O. 1990. Minor cucurbitane-glycosides from fruits of Siraitia grosvenori (Cucurbitaceae). Chem Pharm Bull. 38:2030–2032.
- Nie R. 1994. The decadal progress of triterpene saponins from Cucurbitaceae (1982—1992). Acta Botanica Yunnanica. 16:201–208.
- Pawar RS, Krynitsky AJ, Rader JI. 2013. Sweeteners from plants-with emphasis on Stevia rebaudiana (Bertoni) and Siraitia grosvenorii (Swingle). Anal Bioanal Chem. 405:4397–4407.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yan X, Rivero-Huguet ME, Hughes BH, Marshall WD. 2008. Isolation of the sweet components from Siraitia grosvenorii. Food Chem. 107:1022–1028.