Abstract
The complete chloroplast genome of Leucosceptrum canum, a monotypic species of Lamiaceae with important economic value is reported. The chloroplast of it is 152,739 bp, including a pair of inverted repeats (25,629 bp) regions separated by a large single-copy (83,968 bp) region and a small single-copy (17,513 bp) region. In total, there are 126 genes, 82 protein-coding genes (CDS), 36 tRNAs, and eight rRNAs are annotated in the plastome, in which 111 genes, 77 CDSs, 39 tRNAs, and four rRNAs are unique, respectively. The phylogenetic tree based on 43 complete chloroplast of Lamiaceae comfirmed its delimitation in tribe Pogostemoneae.
Leucosceptrum canum Smith is a small tree belonging to tribe Pogostemoneae of the family Lamiaceae and mainly distributed in the temperate Himalayans, Burma, China, and Nepal (Wu Citation2001; Hansen et al. Citation2007). The species has important economic values, it is an honey plant and is used as an insecticidal agent (Choudhary et al. Citation2004), rarely attacked by herbivores and pathogens (Luo et al. Citation2013). The species is a monotypic genus in Leucosceptrum (Li and Hedge Citation1994). To better understanding systematics of the species and provide scientific basis for the rational utilization and development of L. canum resource, we sequenced and assembled the complete chloroplast genome of it for the first time.
Fresh leaves of L. canum were collected from Kunming Botanic Garden (Yunnan, China). The voucher specimens of L. canum were deposit in the Herbarium of Yunnan Normal University (accession number: CX 187023). Genomic DNA was isolated using a modified CTAB method (Porebski et al. Citation1997) and then fragmented and used to construct short-insert libraries (300 bp) following the manufacture’s protocol (Illumina Inc., USA), and then paired-end sequenced on an Illumina Hiseq X Ten sequencer. The filtered reads (ca. 31.4 million) were assembled using the program NOVOPlasty v2.7.2 (Dierckxsens et al. Citation2016) with complete chloroplast genome of its close relative Pogostemon stellatus (Lour.) Kuntze as reference (GenBank accession No: NC_031434). The assembled chloroplast genome was annotated using GeSeq (Tillich et al. Citation2017).
The complete chloroplast genome of L. canum is assembled to 152,739 bp in length with an average coverage of 594 and GC content of 38.3% (GenBank accession MK681767). The assembled genome containing a large single copy (LSC) region of 83,968 bp, a small single copy (SSC) region of 17,513 bp, and a pair of inverted repeats (IRs) regions of 25,629 bp. In total, there are 126 genes, 82 protein-coding genes (CDS), 36 tRNAs, and eight rRNAs are annotated in the plastome, with 111 unique genes, 77 unique CDSs, 39 unique tRNAs, and four unique rRNAs.
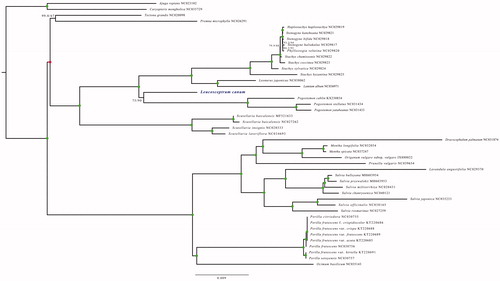
To further determine the phylogenetic position of L. canum with the relative genus with fully sequenced chloroplast genomes, we downloaded 43 Labiatae species with chroloplast genome from Genbank. The whole chloroplast genome sequence of these species were aligned using MUSCLE (Edgar Citation2004); the maximum-likelihood (ML) tree was constructed using IQ_TREE v1.6.10 (Nguyen et al. Citation2015); best-fitted model according to Bayesian information criterion is TVM + F+R3 using ModelFinder (Kalyaanamoorthy et al. Citation2017); branch supports were tested using ultrafast bootstrap (UFBoot) (Hoang et al. Citation2018) and SH-like approximate likelihood ratio test (SH-Alrt) (Guindon et al. Citation2010) with 10,000 replicates. The phylogenetic tree shown that L. canum is nested in the monophyletic clade of tribe Pogostemoneae, confirmed the delimitation of L. canum in tribe Pogostemoneae. Moreover, other topological structures shown by our phylogenetic tree generally in agreement with previously phylogenetic studies of Lamiaceae (Bendiksby et al. Citation2011) ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bendiksby M, Thorbek L, Scheen A, Lindqvist C, Ryding O. 2011. An updated phylogeny and classification of Lamiaceae subfamily Lamioideae. Taxon. 60:471–484.
- Choudhary MI, Ranjit R, Atta-ur-Rahman, Hussain S, Devkota KP, Shrestha TM, Parvez M. 2004. Novel sesterterpenes from Leucosceptrum canum of Nepalese origin. Organic Lett. 6:4139–4142.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Guindon S, Dufayard J, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Hansen DM, Olesen JM, Mione T, Johnson SD, Muller CB. 2007. Coloured nectar: distribution, ecology, and evolution of an enigmatic floral trait. Biol Rev Camb Philos Soc. 82:83–111.
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35:518–522.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589.
- Li H, Hedge IC. 1994. Lamiaceae. Vol. 17. Flora of China. Beijing; St. Louis: Science Press; Missouri Botanical Garden Press.
- Luo S, Hua J, Niu X, Liu Y, Li C, Zhou Y, Jing S, Zhao X, Li S. 2013. Defense sesterterpenoid lactones from Leucosceptrum canum. Phytochemistry. 86:29–35.
- Nguyen L, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15:8–15.
- Tillich M, Lehwark P, Pellizzer T, Ulbrichtjones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Wu Z. 2001. The floristic characteristics and its significance in conservation of semi-humid evergreen broad-leaved forests in Mt Wuliangshan. Acta Bot Yunn. 23:278–286.