Abstract
Rosa rugosa Thunb. is an endangered species in China and also an invasive species in Northern Europe with important ornamental value, economic value, and breeding value, especially wild R. rugosa. In this study, the complete plastid genome of R. rugosa was sequenced and analyzed using next-generation DNA sequencing technology. We subjected the genomic analyses and constructed the maternal phylogenetic relationships. The complete plastid genome was 157,110 bp in length, including a large single copy (LSC) region of 86,214 bp, a small single copy (SSC) region of 18,820 bp and a pair of inverted repeat (IR) regions of 26,038 bp. In total of 116 unique genes were identified, containing 79 protein-coding genes (PCGs), 30 transfer RNA genes, four ribosomal RNA genes, and three pseudogenes (ψycf1, ψycf15, ψycf68), respectively. The phylogenetic analysis revealed that R. rugosa and R. praelucens formed an independent clade with a 100% bootstrap support.
Rosa rugosa Thunb. (‘Mei Gui’ in Chinese) is a shrub plant of the Rosaceae family, native to eastern Asia, includes northeastern China, the Korean Peninsula, Japan, and the Russian Far East (Ku and Robertson Citation2003; Bruun Citation2005). Rosa rugosa is an important germplasm resource for modern roses breeding with many excellent characters, such as unique scent, and high resistance to cold, drought, and disease. It is also an important medicinal and edible plant in China with high economic values and great market potential, especially the rose essential oil. Due to the rapid decline of the number of populations, wild R. rugosa has been listed as a Class II protected plant in China (Fu Citation1992). However, it has been classified as an invasive species in some habitats, particularly in seashores of Northern Europe (Invasive Species Compendium, CABI). Here, we reported the complete plastid genome of R. rugosa (GenBank Accession Number: MK641521) which was characterized based on the Illumina next-generation sequencing techniques. This information is of great significance for the development of scientific strategies to protect and manage the genetic diversity of this plant and the breeding of the genus Rosa.
Samples of R. rugosa were collected from Liaoning province (China; 40°14′04″N, 122°05′12″E). Voucher specimen was deposited in the Herbarium of Kunming Institute of Botany of CAS (KUN). We extracted the total plastid DNA of R. rugosa from young leaf tissue with modified CTAB method (Doyle and Doyle Citation1987). The extracted DNA after purified was sequenced on the Illumina Hiseq Platform (Illumina, San Diego, CA, USA) at Biomarket Technologies Co, LTD (Beijing, China). Then, the high-quality reads were assembled using CLC Genomic Workbench v10 (CLC Bio., Aarhus, Denmark) with the default parameters. These assembled contigs were checked against all the published plastid genomes of the Rosa species by performing BLAST (https://blast.ncbi.nlm.nih.gov/). Plastid contigs were selected, ordered, and oriented according to the reference plastome of R. chinensis var. spontanea (MG523859); the generated gaps between contigs were examined by mapping raw reads to the self-sequence. The complete plastid genome was constructed using Geneious v8.0.2 (Biomatters Ltd., Auckland, New Zealand) and automatically annotated using DOGMA (http://dogma.ccbb.utexas.edu/). To investigate the phylogenetic position of R. rugosa, a total of 16 complete plastid genomes of Rosaceae species were aligned using the online program MAFFT (https://mafft.cbrc.jp/alignment/server/index/index.html), and the maximum-likelihood (ML) phylogenetic tree was then conducted by MEGA v7.0 with 1000 rapid bootstrap replicates (Kumar et al. Citation2016).
Structural analysis of the complete plastid genome of R. rugosa exhibits a typical quadripartite circular structure with 157,110 bp in length. It contained an LSC region of 86,214 bp and an SSC region of 18,820 bp, which were separated by two IR regions of 26,038 bp. The genome encoded 116 unique genes, including 79 PCG genes, 30 tRNA genes, four rRNA genes, and three pseudogenes (ψycf1, ψycf15, ψycf68). Nineteen genes were found to be duplicated in IR regions. Fourteen genes contained one intron and two genes contained two introns. The overall GC content of R. rugosa plastid genome was 37.2%. Furthermore, the GC content of LSC and SSC region was always lower than that of IR regions in all sequenced plastid genomes.
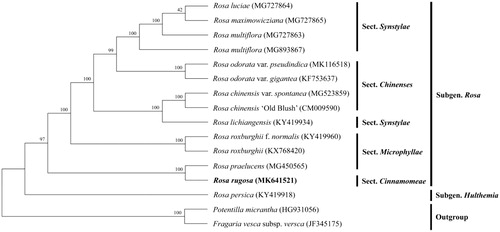
In this study, R. rugosa and thirteen published complete plastid genomes of the genus Rosa (come from NCBI) were used to construct a phylogeny tree, using Potentilla micrantha (HG931056) and Fragaria vesca subsp. vesca (JF345175) in Rosaceae as outgroups. The phylogenetic analysis showed that R. rugosa and R. praelucens (sect. Cinnamomeae) formed an independent clade with a 100% bootstrap support. In addition, sect. Chinenses and sect. Synstylae were closely related with strong support, which was consistent with the former results (Jian et al. Citation2018) ().
Acknowledgements
We appreciate the Laboratory of Molecular Biology in the Germplasm Bank of Wild Species in Southwest China, Kunming Institute of Botany, Chinese Academy of Sciences for providing experimental platform.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bruun HH. 2005. Rosa rugosa Thunb. ex Murray. J Ecol. 93:441–470.
- Centre of Agriculture and Biosciences International (CABI). 2018. Rosa rugosa. In: Invasive species compendium. Wallingford (UK): CAB International. www.cabi.org/isc.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fu LG. 1992. China plant red data book: rare and endangered plants. Beijing: Science Press.
- Jian HY, Zhang YH, Yan HJ, Qiu XQ, Wang QG, Li SB, Zhang SD. 2018. The complete chloroplast genome of a key ancestor of modern roses, Rosa chinensis var. spontanea, and a comparison with congeneric species. Molecules. 23:389–401.
- Ku TC, Robertson KR. 2003. Rosa. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 9. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press; p. 296–339.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.