Abstract
We determined the complete mitochondrial genome of the tree frog, Polypedates braueri using next generation sequencing (NGS) and Sanger sequencing. The mitogenome of P. braueri was 19,904 bp in length, which contained 12 protein-coding genes, 22 tRNAs, two rRNAs, and two control regions (D-Loop). A noncoding sequence (NC) was discovered between tRNALys and ATP6 gene, as well as replaced the original position of ATP8 gene. The ND5 gene was found between the two control regions. More mitochondrial genomic information will contribute to revealing the phylogenetic relationships among species of the genus Polypedates.
The white-lipped tree frog, Polypedates braueri (Anura: Rhacophoridae), is widely distributed in south China (AmphibianChina 2019). Molecular studies of this species usually based on a single gene fragment to estimate the phylogenetic relationships and taxonomic status of tree frogs (Brown et al. Citation2010; Blair et al. Citation2013; Pan et al. Citation2013). Here, we sequenced the complete mitogenome of P. braueri to identify the gene arrangement and performed phylogenetic analyses to address higher level relationships within anura.
The specimen was collected from the Laoban hill (29°58′38.50″ N, 102°59′37.38″ E, Ya’an, Sichuan, China) and deposited in the College of Animal Sciences and Technology, Sichuan Agricultural University, China. DNA was extracted using Ezup type animal genomic DNA extraction kit (Sangon, Shanghai) followed the operation manual. Sequencing libraries were generated using the TruSeq DNA Sample Preparation Kit (Illumina, USA) and the Template Prep Kit (Pacific Biosciences, USA). The genome sequencing was performed by Personal Biotechnology Company (Shanghai, China) using the Pacific Biosciences platform and the Illumina Miseq platform. LA-PCR was used to amplify two large segments covering duplicated control regions where some reads were not assembled using next generation sequencing. The assembled mitogenome sequence was annotated using the MITOS web server under the genetic code for vertebrates (Bernt et al. Citation2013) and was manually compared with the mitochondrial genomes of Rhacophoridae species from GenBank.
The complete mitochondrial genome of P. braueri (GenBank Number: MK687567) was 19,904 bp in length and contained 12 protein-coding genes (ATP6, COI, COII, COIII, ND1, ND2, ND3, ND4, ND4L, ND5, ND6, and Cytb), two ribosomal RNAs (12S and 16SrRNA), and 22 transfer RNAs genes. The base composition was A (29.7%), C (23.3%), G (15.1%), and T (31.9%) with an AT content of 61.6%. With the exception of ND6 and eight tRNAs, most genes were coded on the H-strand, which was consistent with other typical amphibian mitogenomes. Nine protein-coding genes were initiated by ATG, whereas six protein-coding genes (ND1, ATP6, COIII, ND3, ND4, and Cytb) ended with an incomplete stop codon. The tRNA genes (tRNAThr/tRNALeu (CUN)/tRNAPro/tRNAPhe) arranged as “TLPF” gene cluster. Most tRNAs except tRNACys and tRNASer(AGY) could be folded into the perfect cloverleaf secondary structure. A noncoding sequence (NC) was found between tRNALys and ATP6 gene which may have replaced the original position of ATP8 as that of the hypothetical of the previous study (Zhang et al. Citation2005). Two control regions which located on both sides of ND5 were 1,753 bp and 2,757 bp in length, respectively.
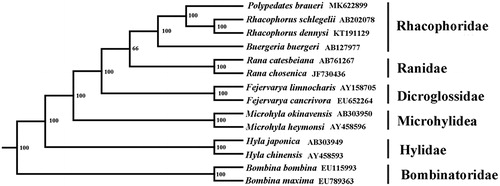
Based on the concatenated nucleotide sequences of 12 protein-coding genes and two rRNAs, the phylogenetic relationships of the P. braueri and the other 13 frogs were reconstructed by MEGA6.0 using maximum-likelihood (ML) method with 1000 bootstrap replications (Tamura et al. Citation2013). Polypedates braueri and the other three Rhacophoridae species were recovered as monophyletic with strong bootstrap value (). Due to the molecular evidence inferred in this study is limited, more mitochondrial genomic information of other tree frogs is necessary in order to elucidate the evolutionary relationships within major lineages of Rhacophoridae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Blair C, Davy CM, Ngo A, Orlov NL, Shi HT, Lu SQ, Gao L, Rao DQ, Murphy RW. 2013. Genealogy and demographic history of a widespread amphibian throughout Indochina. J Hered. 104:72.
- Brown RM, Linkem CW, Siler CD, Sukumaran J, Esselstyn JA, Diesmos AC, Iskandar DT, Bickford D, Evans BJ, McGuire JA, et al. 2010. Phylogeography and historical demography of Polypedates leucomystax in the islands of Indonesia and the Philippines: evidence for recent human-mediated range expansion? Mol Phylogenet Evol. 57:598–619.
- Pan S, Dang N, Wang J, Zheng Y, Rao D, Jiatang LI. 2013. Molecular phylogeny supports the validity of Polypedates impresus Yang 2008. Asian Herpetol Res. 4:124–133.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- The Database of Chinese Amphibians. 2019. Electronic Database accessible at http://www.amphibiachina.org/. Kunming Institute of Zoology (CAS), Kunming, Yunnan, China.
- Zhang P, Zhou H, Liang D, Liu YF, Chen YQ, Qu LH. 2005. The complete mitochondrial genome of a tree frog, Polypedates megacephalus (Amphibia: Anura: Rhacophoridae), and a novel gene organization in living amphibians. Gene. 346:133–143.