Abstract
Coptis omeiensis, as an important Chinese medicinal plant, has been endangered because of overharvesting. To better understand and protect this species, we report the first complete chloroplast genome sequence of C. omeiensis. The chloroplast genome was found to be 154,684 bp in length, and GC content was 38.2%, with large and small single copy regions (LSC with 85,099 bp and SSC with 17,239 bp in length) separated by two Inverted repeat regions (IRs with 26,173 bp). Meanwhile, the sequence contained 113 genes, including 79 protein-coding, 30 tRNA, and 4 rRNA genes. The phylogeny of Ranunculaceae based on the complete cp genomes of 27 related taxa was also presented in this study.
The goldthread genus Coptis is a small genus with about 15 species in Ranunculaceae and famous for its great pharmaceutical importance (Xiang et al. Citation2016, Citation2018). Coptis omeiensis (Chen) C. Y. Cheng is restricted to the mountain cliffs in central Sichuan Province, China, at the altitude of 1000–1700 m. Due to its medicinal importance, this species has been harvested for hundreds of years, and its wild population have been shrinking dramatically (Qin et al. Citation2011). In order to better understand and protect this species, we reported and characterized the first complete chloroplast genome of C. omeiensis by the next-generation sequencing technology. Furthermore, a phylogenomic analysis of Ranunculaceae was also presented.
Dried leaves of C. omeiensis was collected from Mt. Emei, Sichuan, China. Voucher specimen (Z.M. Liang 2018015) was deposited in the Herbarium of Beijing Forestry University (BJFC). Total genomic DNA was extracted using a CTAB method (Doyle and Doyle Citation1987). DNA libraries preparation and 2 × 150 bp pair-end sequencing were performed on an Illumina Hi-Seq 4000 platform at Novogene (http://www.novogene.com, China). Chloroplast reads were extracted using published references (Mamut et al. Citation2018; Zhang et al. Citation2018), and then de novo assembly was done by Geneious R11 (Kearse et al. Citation2012). The annotation was performed using Plann (Huang and Cronk Citation2015), and then verified manually. The plastome sequence was deposited in GenBank (accession number: MK600514).
The plastome sequence of C. omeiensis was 154,684 bp in length, containing two single copy regions (LSC with 85,099 bp and SSC with 17,239 bp) separated by two inverted repeat regions (IRs with 26,173 bp). The sequence GC content was 38.2%, with IR regions having higher GC content (43.1%) than LSC (36.4%) and SSC (32.3%). The cp genome contained 113 genes, including 79 protein-coding, 30 tRNA, and 4 rRNA genes. Among them, 12 protein-coding (atpF, ndhA, ndhB, petB, petD, rpl2, rpoC1, rps16, rpl16) and 6 tRNA genes (trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC) contained a single intron, while three protein-coding genes (clpP, ycf3, rps12) possessed two introns. Of these, the gene rps12 was a trans-spliced gene.
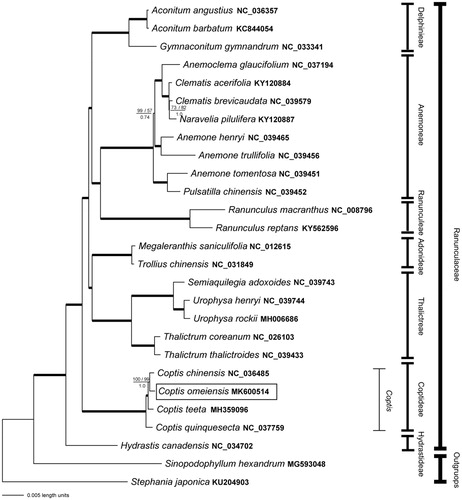
Complete cp genome sequences of other 23 Ranunculaceae species and three outgroup taxa were mined from GenBank (). The Bayesian inference (BI), maximum-likelihood (ML) and parsimony (MP) analyses were conducted for phylogeny reconstruction. BI analysis was performed in MrBayes 3.2.3 (Ronquist et al. Citation2012), with ngen =20,000,000, samplefreq =5000. ML analysis was implemented in RAxML-HPC 8.2.10 (Stamatakis Citation2014) on CIPRES cluster (https://www.phylo.org/), employing the GTR + G model and 1000 bootstrap replicates. MP analysis was carried out in PAUP* 4.0b10 (Swofford Citation2003), with 2000 replicates of random addition and 1000 bootstrap replicates. The phylogenetic framework was consistent with the previous phylogenetic studies on Ranunculaceae (Cossard et al. Citation2016; Liu et al. Citation2018; Mamut et al. Citation2018; Zhang et al. Citation2018; Zhai et al. Citation2019). Subfamily Coptidoideae diverged early from the family. Within Coptis, C. omeiensis was sister to C. chinensis ().
Figure 1. The plastid phylogenomic tree of Coptis omeiensis as well as other Ranunculaceae species inferred from Bayesian analysis. Branches with 100% PP value, 100 likelihood 100 parsimony bootstrap values are shown in bold lines. Otherwise, MP bootstrap values/ML bootstrap values are shown above and pp values are below each branch.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cossard G, Sannier J, Sauquet H, Damerval C, Craene LR, Jabbour F, Nadot S. 2016. Subfamilial and tribal relationships of Ranunculaceae: evidence from eight molecular markers. Plant Syst Evol. 302:419–431.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Pl Sci. 3:1500026.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Liu HJ, He J, Ding CH, Lyu RD, Pei LY, Cheng J, Xie L. 2018. Comparative analysis of complete chloroplast genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica revealing structural variations among genera in tribe Anemoneae (Ranunculaceae). Front Pl Sci. 9:1097.
- Mamut R, Yisilam G, Zhang HW, Hina F, Yang ZP. 2018. The first complete chloroplast genome of Coptis chinensis var. brevisepala, with implication for the phylogeny of Ranunculaceae. Mitochondrial DNA B. 3:953–954.
- Qin JH, Jiang SY, Feng CQ, Zhou Y. 2011. Study of present situation of mild Coptis omeiensis resources and protection approach. J Chengdu Univ. 30:298–301.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Swofford DL. 2003. Paup*: phylogenetic analysis using parsimony (*and other methods), version 4.0 beta version. Sunderland: Sinauer and Assoc.
- Xiang KL, Erst AS, Xiang XG, Jabbour F, Wang W. 2018. Biogeography of Coptis Salisb. (Ranunculales, Ranunculaceae, Coptidoideae), an eastern Asian and north American genus. BMC Evol Biol. 18:74
- Xiang K-L, Wu S-D, Yu S-X, Liu Y, Jabbour F, Erst AS, Zhao L, Wang W, Chen Z-D. 2016. The first comprehensive phylogeny of Coptis (Ranunculaceae) and its implications for character evolution and classification. PLoS One. 11:e0153127
- Zhai W, Duan XS, Zhang R, Guo C, Li L, Xu GX, Shan HY, Kong HZ, Ren Y. 2019. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol Phylogenet Evol. 135:12–21.
- Zhang YY, Dong JX, Feng Y, Wang ZS, Li P. 2018. The first complete chloroplast genome of Coptis quinquesecta, a critically endangered medicinal plant in China. Mitochondrial DNA B. 3:370–372.
