Abstract
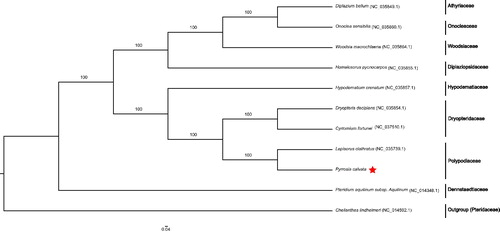
We reported the complete chloroplast genome of Pyrrosia calvata, a traditional Chinese medicinal fern only restricted to Guangxi, China. Its genome is 158,201 bp in length with 41.59% GC content and includes two inverted repeats (IRs) of 26,983 bp separated by LSC and SSC of 82,517 bp and 21,718 bp, respectively. A total of 131 genes are predicted, including 88 protein-coding genes, 34 tRNA genes, eight rRNA genes, and one pseudogene. ML tree indicates that P. calvata is sister to Lepisorus clathratu. This study provides a new perspective to explore the phylogenetic relationship of Pyrrosia.
Pyrrosia calvata (Baker) Ching is a rare epiphytic fern on trunks or rocks belonging to Polypodiaceae. It is 25–70 cm tall with long and deep green blade monomorphic frond. As a drought tolerant fern, P. calvata prefers to direct sun and strong wind occurring on an upper cracking cliff at an altitude of 400–1800 m (Zhang et al. Citation2013). Basically, the fern is a challenge species from limestone area of Guangxi, Southern China, only where its whole plant is used in traditional Chinese medicine as Shiwei (FOLIUM PYRROSIAE) to promote hemostasis and diuresis (Editorial Office of National Chinese Herbal Medicine Collection Citation1975; The Guangxi Zhuang Autonomous Region Health Department Citation1990). Pyrrosia s.l. is a vital terrestrial fern genus in Polypodiaceae, comprising ca. 60 species (Wei et al. Citation2017). However, the infrageneric classification of Pyrrosia is controversial based on chloroplast markers and morphology (Hovenkamp Citation1986; Vasques et al. Citation2017). Furthermore, there is no clear phylogenetic relationship among Pyrrosia, Drymoglossum, and Saxiglossum (Ravensberg and Hennipman Citation1986). Hence, the acquirement of P. calvata chloroplast genome sequence will be conducive to reduce the abuse of medical varieties and solve phylogenetic disputes.
Fresh young leaves of P. calvata were sampled from the South China Botanical Garden, Chinese Academy of Sciences (N23°11′21.66″, E113°21′31.41″) and deposited at the Herbarium of Sun Yat-sen University (SYS; voucher: SS Liu 201620). Its total genomic DNA was extracted using Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China) and further manufactured to average 300 bp paired-end (PE) Illumina genomic library. The whole genome was sequenced on the Illumina Hiseq 2500 platform (Illumina Inc., San Diego, CA, USA). Approximately, 2.28 G high-quality filtered clean data was acquired through Trimmomatic v0.32 (Bolger et al. Citation2014) and further De novo assembled into a complete chloroplast genome using Velvet v1.2.07 (Zerbino and Birney Citation2008). Gene annotation was conducted using DOGMA (Wyman et al. Citation2004) and tRNAscan-SE (Schattner et al. Citation2005) and validated through BLASTX and BLASTN searches against other fern chloroplast genomes. A maximum likelihood (ML) phylogenetic tree was constructed through RAxML v.8.0 (Stamatakis Citation2014) from alignments created with MAFFT (Katoh and Standley Citation2013) using 11 selected complete chloroplast genome sequences of ferns including Cheilanthes lindheimeri as an outgroup. The analysis was performed using 1000 bootstrap replicates.
The complete chloroplast genome of P. calvata is 158,201 bp in length (Genbank accession number: MK679765) with 41.59% GC content. It exhibits typical quadripartite structure with a large single-copy (LSC) region of 82,517 bp, a small single-copy (SSC) region of 21,718 bp and two inverted repeat (IRs) regions of 26,983 bp each. The plastid genome contains a total of 131 genes, including 88 protein-coding genes (PCGs), 34 tRNA genes, eight rRNA genes, and one pseudogene (ndhB). Among them, 14 occur in double copies, involving four PCGs (rps12, rps7, psbA, and ycf2), six tRNA genes (trnN-GUU, trnH-GUG, trnI-GAU, trnA-UGC, trnT-UGU, and trnR-ACG), and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23). Eighteen genes including 12 PCGs and six tRNAs possess one or two introns. ML tree indicates that P. calvata is sister to Lepisorus clathratus with strong bootstrap support (). The chloroplast genome of P. calvata provides a new perspective to explore the phylogenetic relationship of Pyrrosia.
Disclosure statement
The authors declare no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Editorial Office of National Chinese Herbal Medicine Collection. 1975. Quan Guo Zhong Cao Yao Hui Bian [Collection of National Chinese Herbal Medicine]. Beijing: People’s Medical Publishing House. (Chinese).
- Hovenkamp PH. 1986. A monograph of the fern genus Pyrrosia: Polypodiaceae. Vol. 9. Brill Archive; p. 1–280.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Ravensberg W, Hennipman E. 1986. The Pyrrosia species formerly referred to Drymoglossum and Saxiglossum (Filicales, Polypodiaceae.). Leiden Botanical Series. 9:281–310.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- The Guangxi Zhuang Autonomous Region Health Department. 1990. Guangxi Zhong Yao Cai Biao Zhun [Guangxi Chinese medicine standard]. Nanning: Guangxi Science and Technology Publishing House; p. 184–185. (Chinese).
- Vasques DT, Ebihara A, Ito M. 2017. The felt fern genus Pyrrosia Mirbel (Polypodiaceae): a new subgeneric classification with a molecular phylogenic analysis based on three plastid markers. Acta Phytotax Geobot. 68:65–82.
- Wei XP, Qi YD, Zhang XC, Luo L, Shang H, Wei R, Liu HT, Zhang BG. 2017. Phylogeny, historical biogeography and characters evolution of the drought resistant fern Pyrrosia Mirbel (Polypodiaceae) inferred from plastid and nuclear markers. Sci Rep. 7:12757.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang XC, Lu SG, Lin YX, Qi XP, Moore S, Xing FW, Wang FG, Hovenkamp PH, Gilbert MG, Nooteboom HP, et al. 2013. Polypodiaceae. In: Wu ZY, Raven PH, Hong DY, eds., Flora of China. Vol. 2–3 (Pteridophytes). Beijing: Science Press; p. 758–850.