Abstract
In the present study, we presented the complete mitochondrial genome of a plant pathogen, Plectosphaerella sp. It has a total length of 31,004 bp with the base composition as follows: A (34.66%), T (36.76%), C (13.48%), and G (15.10%). The mitogenome contains 20 protein-coding genes, 2 ribosomal RNA genes (rRNA), and 28 transfer RNA (tRNA) genes. The taxonomic status of the Plectosphaerella sp. mitogenome exhibited a close relationship with the Verticillium genus.
Plectosphaerella is a genus of filamentous fungi belonging to Glomerellales, Ascomycota (Su et al. Citation2017). Plectosphaerella spp. are widely distributed on the earth and are well-known as plant pathogens (Pétriacq et al. Citation2016). Plectosphaerella cucumerina has been implicated as the causal pathogen of tomato wilt (Xu et al. Citation2014), lettuce necrotic leaf spot (Carrieri et al. Citation2014), cabbage root rot (Li et al. Citation2017), and endive leaf spot (Garibaldi et al. Citation2013), which leads to large losses of crops. In addition, some Plectosphaerella species can be used as biological control agents against potato cyst nematodes (PCN) and other nematode species (Atkins et al. Citation2003). Plectosphaerella cucumerina also showed anti-biofilm and antivirulence activities against Pseudomonas aeruginosa (Zhou et al. Citation2017). To the best of our knowledge, this is the first report of the complete mitochondrial genome from the Plectosphaerella genus, which will provide a reference for understanding the phylogeny and evolution of this important genus.
The specimen (P. cucumerina sp.) was isolated from the tomato-growing soil in Chengdu, Sichuan, China (106.77 E; 30.39 N) and was stored in Sichuan Academy of Agricultural Sciences (No. Pcua1). The total genomic DNA of P. cucumerina sp. was extracted using Fungal DNA Kit D3390-00 (Omega Bio-Tek, Norcross, GA, USA) and purified through a Gel Extraction Kit (Omega Bio-Tek, Norcross, GA, USA). Purified DNA was stored in the sequencing company (BGI Tech, Shenzhen, China). Sequencing libraries were constructed with purified DNA following the instructions of NEBNext™ Ultra™ II DNA Library Prep Kit (NEB, Beijing, China). Whole genomic sequencing was performed by the Illumina HiSeq 2500 Platform (Illumina, SanDiego, CA, USA). The raw data obtained was first passed through the quality control step. The complete mitochondrial genome was de novo assembled as implemented by SPAdes 3.9.0 (Bankevich et al. Citation2012). Gaps among contigs were filled by using MITObim V1.9 (Oslo, Norway) (Hahn et al. Citation2013). The determined mitogenome was annotated using the MFannot tool (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl), combined with manual corrections. tRNAs were annotated by tRNAscan-SE (Lowe and Eddy, Citation1997).
The total length of P. cucumerina sp. circular mitogenome is 31,004 bp. This mitogenome was submitted to GenBank database under accession No. MK697668. The circular mitogenome contains 20 protein-coding genes, 2 ribosomal RNA genes (rns and rnl), and 28 transfer RNA (tRNA) genes. The base composition of the genome is as follows: A (34.66%), T (36.76%), C (13.48%), and G (15.10%).
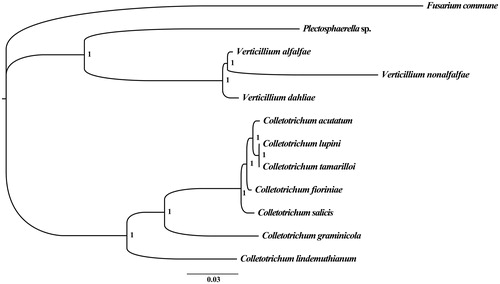
To validate the phylogenetic position of P. cucumerina sp., we constructed phylogenetic trees of 12 closely related species from Hypocreomycetidae. Bayesian analysis (BI) were used to construct the phylogenetic trees with the 14 core protein-coding genes and 2 rRNA genes according to Qiang, Min et al. Citation2018a; Qiang, Qiangfeng et al. Citation2018b. As shown in the phylogenetic tree (), the taxonomic status of the P. cucumerina sp. based on mitogenome exhibited a close relationship with the Verticillium genus.
Figure 1. Molecular phylogenies of 12 species based on Bayesian inference analysis of the combined mitochondrial gene set (14 core protein-coding genes + two rRNA genes). Node support values are Bayesian posterior probabilities (BPP). Mitogenome accession numbers used in this phylogeny analysis: Fusarium commune (NC_036106), Colletotrichum acutatum (NC_027280), Colletotrichum fioriniae (NC_030052), Colletotrichum graminicola M1.001 (NW_007361658), Colletotrichum lindemuthianum (NC_023540), Colletotrichum lupini (NC_029213), Colletotrichum salicis (NC_035496), Colletotrichum tamarilloi (NC_029706), Verticillium alfalfae VaMs.102 (NW_003315039), Verticillium dahliae (NC_008248), Verticillium nonalfalfae (NC_029238).
Disclosure statement
The authors have declared that no competing interests exist.
Additional information
Funding
References
- Atkins SD, Clark IM, Sosnowska D, Hirsch PR, Kerry BR. 2003. Detection and quantification of Plectosphaerella cucumerina, a potential biological control agent of potato cyst nematodes, by using conventional PCR, real-time PCR, selective media, and baiting. Appl Environ Microbiol. 69:4788–4793.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Carrieri R, Pizzolongo G, Carotenuto G, Tarantino P, Lahoz E. 2014. First report of necrotic leaf spot caused by Plectosphaerella cucumerina on lamb's lettuce in southern italy. Plant Dis. 98:998.
- Garibaldi A, Gilardi G, Ortu G, Gullino ML. 2013. First report of a new leaf spot caused by Plectosphaerella cucumerina on field grown endive (cichorium endivia) in Italy. Plant Dis. 97:848–848.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Li PL, Chai AL, Shi YX, Xie XW, Li BJ. 2017. First report of root rot caused by Plectosphaerella cucumerina on cabbage in China. Mycobiology. 45:110–113.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Pétriacq P, Stassen JH, Ton J. 2016. Spore density determines infection strategy by the plant pathogenic fungus Plectosphaerella cucumerina. Plant Physiol. 170:2325–2339.
- Qiang L, Min L, Mei Y, Chuan X, Xin J, Zuqin C, Wenli H. 2018. Characterization of the mitochondrial genomes of three species in the ectomycorrhizal genus Cantharellus and phylogeny of Agaricomycetes. Int J Biol Macromol. 118:756–769.
- Qiang L, Qiangfeng W, Cheng C, Xin J, Zuqin C, Chuan X, Ping L, Jian Z, Wenli H. 2018. Characterization and comparative mitogenomic analysis of six newly sequenced mitochondrial genomes from ectomycorrhizal fungi (Russula) and phylogenetic analysis of the Agaricomycetes. Int J Biol Macromol. 119:792–802.
- Su L, Deng H, Niu YC. 2017. Phylogenetic analysis of Plectosphaerella species based on multi-locus DNA sequences and description of P. sinensis sp. nov. Mycol Progress. 16:823–829.
- Xu J, Xu XD, Cao YY, Zhang WM. 2014. First report of greenhouse tomato wilt caused by Plectosphaerella cucumerina in china. Plant Dis. 98:158.
- Zhou J, Bi S, Chen H, Chen T, Yang R, Li M, Fu Y, Jia AQ. 2017. Anti-biofilm and antivirulence activities of metabolites from Plectosphaerella cucumerina against Pseudomonas aeruginosa. Front Microbiol. 3:769.
