Abstract
The relationship of Matrona and Atrocalopteryx (Odonata: Calopterygidae) is still unclear. To better understand the phylogenetic relationship of Matrona and Atrocalopteryx, we sequenced and annotated two complete mitochondrial genomes of Matrona basilaris sampled from two different locations. The length of the two complete mitochondrial genomes of M. basilaris is 16,149 bp and 15,893 bp for the specimens collected in Jinhua, Zhejiang Province and Tianmushan, Zhejiang Province, China, respectively. The two mitochondrial genomes include the typical invertebrate set of 37 genes: 13 protein-coding genes (PGCs), 22 tRNA genes, and 2 rRNA genes. The nucleotide composition of the mitogenome is similar to other odonates with high content of A + T (68.9%) and all PCGs use ATN as the start codon. Tandem repeats were detected in the control regions of the two M. basilaris samples that accounted for the different sequence lengths of the mitochondrial genomes from the two locations. Finally, BI and ML phylogenetic analysis based on the concatenated nucleotide sequences of the 13 PCGs supported the conclusion that M. basilaris is a sister clade to Atrocalopteryx melli.
The relationship between Matrona and Atrocalopteryx (Odonata: Calopterygidae) is unclear (Dumont et al. Citation2007; Guan et al. Citation2012). To help resolve this, we sequenced the mitochondrial genome of two specimens of Matrona basilaris (Zygoptera: Calopterygidae) to better determine their phylogenetic relationship within Odonata. Two samples of M. basilaris were collected from Jinhua (JH), Zhejiang province and Tianmushan (TMS), Zhejiang province, China, respectively. Using an Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech Company, Shanghai, China), the total genomic DNA was isolated from the thorax muscle of the two samples. Remaining samples and DNA extracts were stored in the lab of Dr. Zhang, College of Chemistry and Life Science, Zhejiang Normal University. A set of conserved and modified primers (Simon et al. Citation2006; Zhang et al. Citation2008; Zhang, Cai, et al. Citation2018; Zhang, Yu, et al. Citation2018) and six specific primers were designed for polymerase chain reaction (PCR) amplification. The sequences of the two mitogenomes are deposited in GenBank with accession numbers MK722304 and MK722305 for JH and TMS, respectively.
The two complete mitochondrial genomes of M. basilaris are 16,149 bp (JH) and 15,893 bp (TMS) in length, respectively. All protein-coding genes of the two samples are AT-biased (68.9%), with atp8 showing the highest A + T content (76.8% and 76.1%) whereas cox1 has the lowest A + T content (both samples are 63%); these results are similar to other odonates (Lin et al. Citation2010; Lorenzo-Carballa et al. Citation2014; Feindt et al. Citation2016). All protein-coding genes of M. basilaris employ the characteristic invertebrate specific mitochondrial start codons: cox2, cox3, atp6, nad4, nad4L, cytb and nad1 start with ATG; cox1 and nad3 with ATA; atp8 and nad5 with ATT; and nad2 and nad6 with ATC. Eight protein-coding genes have the standard stop codons TAA (cytb, nad2, nad6, nad4L, atp6, atp8 and cox1) and TAG (nad1) whereas cox2, cox3, nad3 and nad5 have incomplete stop codons of a single T, and nad4 has an incomplete TA stop codon. Tandem repeats occured in the control region and were responsible for the different full lengths of the M. basilaris from the JH and TMS locations.
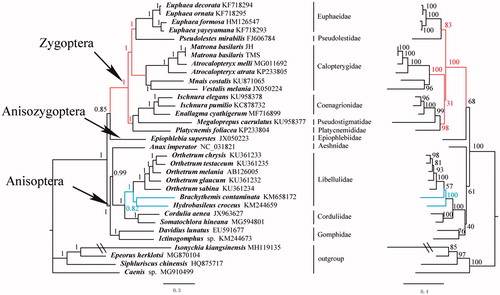
We constructed Bayesian inference (BI) and maximum likelihood (ML) trees from the 13 PCGs of M. basilaris and 28 other Odonata species (Yamauchi et al. Citation2004; Lin et al. Citation2010; Lorenzo-Carballa et al. Citation2014; Tang et al. Citation2014; Feindt et al. Citation2016; Yong et al. Citation2016; Yu et al. Citation2016; Zhang et al. Citation2017) and using four outgroups: Isonychia kiangsinensis (Ye et al. Citation2018), Epeorus herklotsi (Gao et al. Citation2018), Siphluriscus chinensis (Li et al. Citation2014) and Caenis sp. (Cai et al. Citation2018) (). In order to select conserved regions of the nucleotide sequences, each alignment was performed by Gblock 0.91b (Castresana Citation2000) using default settings. BI and ML analyses were performed by MrBayes 3.1.2 (Huelsenbeck and Ronquist Citation2001) and RAxML 8.2.0 (Stamatakis Citation2014), respectively. Both BI and ML phylogenetic trees showed that M. basilaris is a sister clade to Atrocalopteryx melli. However, the monophyly of Atrocalopteryx failed because M. basilaris is clustered into Atrocalopteryx.
Figure 1. Phylogenetic tree of the relationships among 29 species of Odonata including Matrona basilaris, based on the nucleotide dataset of the 13 mitochondrial protein-coding genes. Four outgroups (Isonychia kiangsinensis, Epeorus herklotsi, Siphluriscus chinensis and Caenis sp.) were used. Numbers above branches specify posterior probabilities from Bayesian inference (BI) (left) and bootstrap percentages from maximum likelihood (ML) (right) analyses. The red and blue lines indicate a different topology between BI and ML analyses. GenBank accession numbers of all species are also shown.
Disclosure statement
The authors report no conflicts of interest and are responsible for the content and writing of the paper.
Additional information
Funding
References
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Cai YY, Gao YJ, Zhang LP, Yu DN, Storey KB, Zhang JY. 2018. The mitochondrial genome of Caenis sp. (Ephemeroptera: Caenidae) and the phylogeny of Ephemeroptera in Pterygota. Mitochondrial DNA B. 3:577–579.
- Dumont HJ, Vierstraete A, Vanfleteren JR. 2007. A revised molecular phylogeny of the Calopteryginae (Zygoptera: Calopterygidae). Odonatologica. 36:365–372.
- Feindt W, Osigus HJ, Herzog R, Mason CE, Hadrys H. 2016. The complete mitochondrial genome of the neotropical helicopter damselfly Megaloprepus caerulatus (Odonata: Zygoptera) assembled from next generation sequencing data. Mitochondrial DNA B. 1:497–499.
- Gao XY, Zhang SS, Zhang LP, Yu DN, Zhang JY, Cheng HY. 2018. The complete mitochondrial genome of Epeorus herklotsi (Ephemeroptera: Heptageniidae) and its phylogeny. Mitochondrial DNA B. 3:303–304.
- Guan Z, Han BP, Vierstraete A, Dumont HJ. 2012. Additions and refinements to the molecular phylogeny of the Calopteryginae S. L. (Zygoptera: Calopterygidae). Odonatologica. 41:17–24.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Li D, Qin JC, Zhou CF. 2014. The phylogeny of Ephemeroptera in Pterygota revealed by the mitochondrial genome of Siphluriscus chinensis (Hexapoda: Insecta). Gene. 545:132–140.
- Lin CP, Chen MY, Huang JP. 2010. The complete mitochondrial genome and phylogenomics of a damselfly, Euphaea formosa support a basal Odonata within the Pterygota. Gene. 468:20–29.
- Lorenzo-Carballa MO, Thompson DJ, Cordero-Rivera A, Watts PC. 2014. Next generation sequencing yields the complete mitochondrial genome of the scarce blue-tailed damselfly, Ischnura pumilio. Mitochondrial DNA. 25:247–248.
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Systemat. 37:545–579.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tang M, Tan M, Meng G, Yang S, Su X, Liu S, Song W, Li Y, Wu Q, Zhang A, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes – a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42:e166
- Yamauchi MM, Miya MU, Nishida M. 2004. Use of a PCR-based approach for sequencing whole mitochondrial genomes of insects: two examples (cockroach and dragonfly) based on the method developed for decapod crustaceans. Insect Mol Biol. 13:435–442.
- Ye QM, Zhang SS, Cai YY, Storey KB, Yu DN, Zhang JY. 2018. The complete mitochondrial genome of Isonychia kiangsinensis (Ephemeroptera: Isonychiidae). Mitochondrial DNA B. 3:541–542.
- Yong HS, Song SL, Suana IW, Eamsobhana P, Lim PE. 2016. Complete mitochondrial genome of Orthetrum dragonflies and molecular phylogeny of Odonata. Biochem Systemat Ecol. 69:124–131.
- Yu PP, Cheng XF, Ma Y, Yu DN, Zhang JY. 2016. The complete mitochondrial genome of Brachythemis contaminata (Odonata: Libellulidae). Mitochondrial DNA A. 27:2.
- Zhang JY, Zhou CF, Gai YH, Song DX, Zhou KY. 2008. The complete mitochondrial genome of Parafronurus youi (Insecta: Ephemeroptera) and phylogenetic position of the Ephemeroptera. Gene. 424:18–24.
- Zhang L, Wang XT, Wen CL, Wang MY, Yang XZ, Yuan ML. 2017. The complete mitochondrial genome of Enallagma cyathigerum (Odonata: Coenagrionidae) and phylogenetic analysis. Mitochondrial DNA B. 2:640–641.
- Zhang LP, Cai YY, Yu DN, Storey KB, Zhang JY. 2018. Gene characteristics of the complete mitochondrial genomes of Paratoxodera polyacantha and Toxodera hauseri (Mantodea: Toxoderidae). PeerJ. 6:e4595.
- Zhang LP, Yu DN, Storey KB, Cheng HY, Zhang JY. 2018. Higher tRNA gene duplication in mitogenomes of praying mantises (Dictyoptera: Mantodea) and the phylogeny within Mantodea. Int J Biol Macromol. 111:787–795.
