Abstract
Moringa oleifera Lam. is a member of the Moringaceae family which includes 13 species. The plant is cultivated throughout tropical and subtropical areas due to its adaptability, also used worldwide as nutritional supplement and traditional medicine. In this study, the complete chloroplast genome (cp) of M. oleifera was sequenced, assembled, and analyzed. Our results indicated that the cp genome of M. oleifera was 160,600 bp in length, containing a large single copy (LSC, 88,727 bp) region and a small single copy (SSC, 18,883 bp) region which separated by two reduced inverted repeats (IRs, 26,495 bp). A total of 108 unique genes were identified, including 74 protein-coding genes, 30 transfer RNA (tRNA) genes, and four ribosomal RNA (rRNA) genes. The overall GC content of M. oleifera cp genome was 36.8%. Most of the 47 simple sequence repeats (SSRs) were mononucleotides motifs of A/T types and were found to be located in non-coding regions. Phylogenetic analysis of M. oleifera showed that M. oleifera was the base group of Rhoeadales with strongly bootstrap supported which is consist with morphological taxonomy.
Moringa oleifera is an evergreen and fast-growing tree, native to north-western India, and gained popularity in certain developing countries for its high-nutrition content and adaptability to arid and semi-arid environments. A wide variety of nutritional and medicinal virtues have been attributed to its roots, bark, leaf, flowers, fruits, and seeds. Leaf of M. oleifera has been used to treat malnutrition. Important medicinal properties of the plant include antipyretic, antiepileptic, antiinflamatory, antiulcerative, antihypertensive, cholesterol lowering, antioxidant, antidiabetic, hepatoprotective, antibacterial, and antifungal activities. As such an important edible plant, its phylogenetic position is still unclear in previous studies because of the lack of sampling. Here, we characterized the complete chloroplast (cp) genome sequence of M. oleifera based on the genome skimming sequencing data, detected the occurrence, type, and distribution of simple sequence repeats (SSRs) and constructed the phylogenetic tree based on the neighbour-joining (NJ) method.
The fresh leaves of M. oleifera were collected from the laboratory of Yunnan Agricultural University. The voucher specimen was deposited at Herbarium, Kunming Institute of Botany, CAS (KUN). Total genomic DNA was isolated from fresh leaves using the modified cetyltrimethyl ammonium bromide (CTAB) method (Doyle Citation1987) to construct chloroplast DNA libraries. The extracted DNA was sequenced using the Illumina Hiseq2000 (Illumina, CA, USA). In all, 187 M of 150-bp raw paired reads were retrieved. Prior to chloroplast de novo assembly, low-quality reads were filtered out, and resultant clean reads were assembled using GetOrganelle pipeline (https://github.com/Kinggerm/GetOrganelle). All the contigs were checked against the reference genome of Papaver somniferum (NC029434) using BLAST (https://blast.ncbi.nlm.nih.gov/), and aligned contigs were oriented according to the reference genome.
The genome was automatically annotated using the CpGAVAS pipeline (Liu et al. Citation2012), and start/stop codons and intron/exon boundaries were adjusted in Geneious 8.1(Kearse et al. Citation2012). The tRNA was identified through tRNAscan-SE v2.0 (Lowe and Chan Citation2016). Sequence data were deposited into GenBank. A physical map of the cp genome was generated using the online tool OGDraw v1.2 (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2007). The complete cp genome sequence was submitted to the GenBank under Accession Number of MK713965.
The complete cp genome of M. oleifera was 160,600 bp in length. It was the typical quadripartite structure, in which two inverted repeats (IRs, 26,495 bp) were separated by the large single-copy (LSC, 88,727 bp) and the small single-copy (SSC, 18,883 bp) regions. The cp genome had 108 genes, including 74 protein-coding genes (PCGs), 30 tRNA genes, and four rRNA genes. Most of genes occurred as single-copy while eight PCGs (ndhB, rpl2, rpl23, rps12, rps19, rps7, ycf1, and ycf2), seven tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and four rRNA genes (rrn16, rrn23, rrn4.5, and rrn5) had two copies. In addition, two PCGs (clpP and ycf3) had two introns each; seven PCGs (atpF, ndhA, ndhB, petB, petD, rpl2, and rpl16) contained one intron. The overall GC content of the cp genome was 36.8%, while that of LSC, SSC, and IR regions was 34.6%, 30.6%, and 42.6%, respectively. Phobos v3.3.12 (Leese et al. Citation2008) and SSRHunter (Li and Wan Citation2005) was used to find the SSR markers present in the chloroplast genome of M. oleifera. It uses a recursive algorithm to search for repeats with lengths between one and six. The unit sizes of mono-, di-, tri-, tetra-, penta-, and hexa-nucleotide repeats were set to minimum number of repeats of 10, 5, 4, 3, 3, and 3, respectively. We removed one inverted repeat region (IRa) in SSRs analysis and manually checked the detected repeats. The occurrence and distribution of different types of SSR in the cp genome are summarized in . A total of 47 SSRs were detected throughout the cp genome of M. oleifera, with 23, 10, 5, 7, and 2 for mono-, di-, tri-, tetra-, and hexa-nucleotide repeats, respectively. The majority of the mono-nucleotides were A or T and all of di-nucleotides were AT or TA repeats, which was consistent with the A/T-richness in the complete cp genome (Xuan et al. Citation2013).
Table 1. Simple sequence repeats (SSRs) identified in the chloroplast genome of M. oleifera.
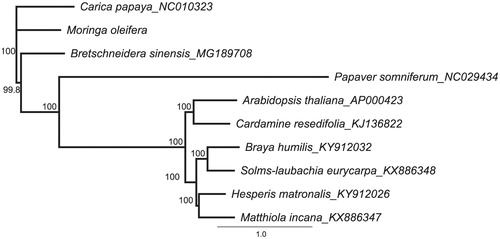
For NJ phylogenetic analysis, the complete chloroplast genome of eight Rhoeadales species and one Parietales species were downloaded from NCBI GenBank. We chose Carica papaya L. of Parietales as an outgroup. The combined datasets based on plastid genomes of 10 species were aligned using MAFFT v7.307 (Katoh and Standley Citation2013). A neighbour-joining (NJ) phylogenetic tree was constructed in Geneious 8.1 (Kearse et al. Citation2012) with the Tamura-Nei genetic distance model, and a total of 1000 bootstrap replicates were performed. The phylogenetic results showed that M. oleifera was the base group of Rhoeadales with strongly bootstrap supported which is consistent with morphological taxonomy ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647.
- Leese F, Mayer C, Held C. 2008. Isolation of microsatellites from unknown genomes using known genomes as enrichment templates. Limnol Oceanogr Methods. 6:412–426.
- Li Q, Wan JM. 2005. SSRHunter: development of a local searching software for SSR sites. Hereditas. 27:808.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13:715.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Xuan Y, Lei G, Bo W, Su YJ, Wang T. 2013. The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): Evolutionary comparison of Cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biol Evol. 5:688–698.