Abstract
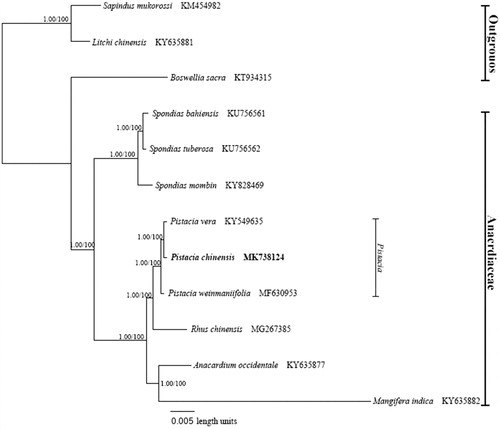
In this study, the complete chloroplast genome of Pistacia chinensis Bunge, a bioenergy tree whose seed oil has been proposed as a promising feedstock for both edible and industrial oil, was generated. The chloroplast genome of P. chinensis was 160,618 bp long, which was comprised of a large single copy (LSC) region of 88,371 bp, a small single copy (SSC) region of 19,057 bp, and a pair of inverted repeats (IRs) of 26,595 bp. The chloroplast genome GC content was 37.87%, contained 132 genes, including 87 protein-coding genes, eight rRNA genes, and 37 tRNA genes. Phylogenetic analysis revealed that P. chinensis was closely grouped with P. vera and P. weinmaniifolia, belonging to the Anacardiaceae family.
Pistacia chinensis Bunge is a kind of woody oil plant, which the oil content of the seeds is normally above 30% and the proportion of unsaturated fatty acid (UFA) is up to 82% (Li et al. Citation2012). Currently, P. chinensis is considered as a good raw material in bio-diesel production for the high content and good quality of its oil (Yin et al. Citation2018). However, previous studies in P. chinensis have mainly focused on its seed oil (Li et al. Citation2018), there have been no reports regarding the genome of this plant species until now. Here, we characterized a chloroplast genome sequence of P. chinensis and analyzed its phylogenetic relationship with other Anacardiaceae species. The genome information can be further applied to conservation genetics, taxonomy, phylogeny, and breeding of this plant species.
The young leaves of P. chinensis were collected from Taiting Village, Qinshui County, Shanxi province, China (N 35°32′9.64″, E 112°06′53.92″). The voucher specimen was deposited in the Herbarium of Beijing Forestry University (BJFC) (under collection numbers of PcB001). The genomic DNAs were extracted using Plant Genomic DNA Kit. These DNAs were manufactured to average 150 bp paired-end (PE) library and sequenced on the Illumina Hiseq X–Ten Sequencing platform at Yuanyi, Beijing, China (http://www.ori–gene.cn/). SOAP de novo and BLAT (Luo et al. Citation2012; Kent Citation2002) were used to assemble the clean reads into a single draft sequence using the closely related species chloroplast genome as reference. The chloroplast sequence was then annotated using CpGAVAS (Liu et al. Citation2012) and DOGMA (Wyman et al. Citation2004) (http://dogma.ccbb.utexas.edu/, China). All annotations were verified by Geneious Prime finally (Kearse et al. Citation2012).
The complete chloroplast genome of P. chinensis (accession number: MK738124) was 160,618 bp in length with 37.87% overall GC content. The genome consisted of four distinct parts, a large single copy (LSC) region of 88,371 bp, a small single copy (SSC) region of 19,057 bp, and a pair of inverted repeats (IRa and IRb) of 26,595 bp. The genome contained 132 genes from which 87 are protein-coding, eight rRNA, and 37 tRNA genes.
Complete chloroplast genome sequence of P. chinensis was subjected to phylogenetic analysis with eight species belonging to the Anacardiaceae family and three outgroups using Maximum Parsimony (MP) method with 1000 bootstrap values in the PAUP* 4.0a (Swofford Citation2003). Bayesian inference (BI) was performed with Mrbayes v3.2.7 (Ronquist et al. Citation2012). The phylogenetic analysis revealed that P. chinensis was closely located with two Pistacia species, P. vera (KY549635) and P. weinmaniifolia (MF630953), belonging to the Anacardiaceae family (), which is coherent with previous reports (Yi et al. Citation2008; AL-Saghir Citation2010).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- AL-Saghir MG. 2010. Phylogenetic analysis of the genus Pistacia L. (Anacardiaceae) based on morphological data. Asian J Plant Sci. 9:28–35.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kent WJ. 2002. BLAT-the BLAST-like alignment tool. Genome Res. 12:656–664.
- Li TF, Wang XQ, Jiao J, Liu JZ, Zhang HX, Niu LL, Zhao CJ, Gu CB, Efferth T, Fu YJ. 2018. Catalytic transesterification of Pistacia chinensis seed oil using HPW immobilized on magnetic composite graphene oxide/cellulose microspheres. Renew Enrg. 127:1017–1025.
- Li X, He XY, Li ZL, Wang YD, Wang CY, Shi H, Wang F. 2012. Enzymatic production of biodiesel from Pistacia chinensis bge seed oil using immobilized lipase. Fuel. 92:89–93.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Liu XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 13:715.
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, et al. 2012. SOAPdenovo2: an empirically improved memory–efcient short–read de novo assembler. Gigascience. 1:18.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (and other methods). Version 4.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yi T, Wen J, Golan‐Goldhirsh A, Parfitt DE. 2008. Phylogenetics and reticulate evolution in Pistacia (Anacardiaceae). Am J Bot. 95:241–251.
- Yin F, Yang XF, Wang HL, Liu L, Meng XP. 2018. Evaluation of the potential land for biofuel plant development in the Shaanxi Province, China. Geol J. 53:332–341.