Abstract
The complete chloroplast genome sequence of Daphne tangutica Maxim. (Thymelaeaceae), a medicinal plant distributed in northwest alpine region of China, was characterized the complete nucleotide sequence of chloroplast (cp) genome. The total length of cp genome is 169,944 bp, including a large single copy (LSC) region of 84,884 bp, a small single copy (SSC) region of 2248 bp, and a pair of inverted repeats (IRs) of 41,406 bp. The cp genome of D. tangutica contains 137 genes, including 90 protein-coding genes (75 PCG species), 39 transfer RNA genes (31 tRNA species), and eight rRNA genes (four RNA species). A total of 12 genes (tRNA-Lys (UUU), tRNA-Leu (UAA), tRNA-Val (UAC), tRNA-Ile (GAU), tRNA-Ala (UGC), rpl16, rpoC1, petD, rps16, rpl2, ndhB, and ndhA) contains a single intron, and two genes (ycf3 and rps12) contains two introns. The GC content in whole cp genome, LSC region, SSC region, and IR region were 36.8%, 34.8%, 28.6%, and 38.9%, respectively, which are similar to other Thymelaeaceae plants. Phylogenetic analysis suggested that D. tangutica has the closely reltionship with congeneric D. kiusiana.
Daphne tangutica Maxim. (Thymelaeaceae), a medicinal plant mainly distributed in west China, has been used as a traditional Tibetan medicine named ‘Shen Xiang Na Ma’ for the treatment of rheumatoid arthritis and apoplexia (Zhou Citation2014; Li et al. Citation2010). Owing to important medicinal value, wild population of D. tangutica has been over-exploited in recent years. In this study, we characterized the complete chloroplast (cp) genome sequence of D. tangutica using high-throughput Illumina sequencing technology, the result will be useful for researching the phylogenetic relationships of D. tangutica and it may help the conservation of this endangered species. The annotated genome sequence has been submitted to GenBank with the accession number MK455880.
Fresh leaves of D. tangutica were collected from Qinlian Mountains (37°56′19.67″N, 100°22′59″E, 3326 m), Gansu, China. The total DNA was extracted using modified CTAB method (Doyle Citation1987) and used for Illumina HiSeq sequencing by Biomarker Technologies, Inc. (Beijing, China), with voucher specimen were deposited at the Library No. 6, Sample Center, Shunjie Building, No. 12, Shunyi District, Beijing (BMK4230-P02D). In total, 21,000,978 of 150-bp raw paired-end reads were obtained and were quality-trimmed using the software in NGSQC Toolkit v2.3.3 (Patel and Jain Citation2012) with 2% allowed ambiguous bases. The sequence of Daphne kiusiana (KY991380) was used as reference for the assemblance of 140,196 individual chloroplast genome reads yielded a mean coverage of 123.4X using MITObim v1.8 (Hahn et al. Citation2013). The cp genome sequence was annotated using GENEIOUS R 8.0.2 (Biomatters Ltd., Auckland, New Zealand). Subsequently, a physical map of the cp genome was generated using the software OGDraw v1.2 (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Lohse et al. Citation2013).
The total length of nucleotide sequence of plastid genome is 169,944 bp, including a large single copy (LSC) region of 84,884 bp, a small single copy (SSC) region of 2248 bp, and a pair of inverted repeats (IRs) of 41,406 bp. The cp genome of D. tangutica contains 137 genes, including 90 protein-coding genes (75 PCG species), 39 transfer RNA genes (31 tRNA species), and eight rRNA genes (four RNA species). A total of 12 genes (tRNA-Lys, tRNA-Leu, tRNA-Val, tRNA-Ile, tRNA-Ala, rpl16, rpoC1, petD, rps16, rpl2, ndhB, and ndhA) contains a single intron, and two genes (ycf3 and rps12) contains two introns. The GC content in whole cp genome, LSC region, SSC region, and IR region were 36.8%, 34.8%, 28.6%, and 38.9%, respectively, which are similar to other Thymelaeaceae plants.
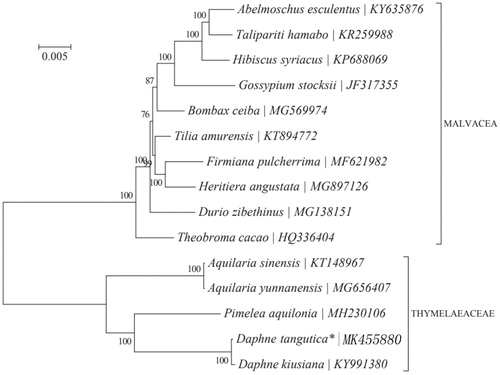
To investigate the phylogenetic position of D. tangutica, a neighbour-joining (NJ) phylogenetic tree () was made based on the concatenated coding sequences of cp PCGs for 14 plastid genomes from published species of Thymelaeaceae using MEGA7 with 1000 bootstrap replicates (Kumar et al. Citation2016) (http://www.megasoftware.net/). The result of the phylogenetic analysis shows that D. tangutica has closely reltionship with congeneric D. kiusiana. The complete cp genome sequence adds valuable information for the study of the genetic diversity of D. kiusiana and Thymelaeaceae.
Disclosure statement
The authors report no conflicts of interest, and are solely responsible for the content and writing of this paper.
Additional information
Funding
References
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Li P, Zhang XF, Deng Y, Zhou Y, Wang H, Ding LS. 2010. Chemical constituents investigation of Daphne tangutica. Fitoterapi. 81:38–41.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucl Acids Res. 41:W575–W581.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Zhou QP. 2014. Botanical illustration of medicinal plants in Qinghai, Volume One. Jiangsu: Jiangsu Science and Technology Press, 156–157.