Abstract
In this study, the complete sequence of the mitochondrial DNA (mtDNA) of Ctenolepisma villosa was obtained using next-generation sequencing approaches and de novo assembly. The molecule was found to be 15,488 bp in length. It is the fourth complete mt genome sequence from the Zygentoma. The mt genomes are circular and encode 37 genes and a large non-coding region. The overall structure (gene number, orientation, and order) of the mt genomes is the same as that found in three other sequenced species. All protein-coding sequences except cox1 start with the typical ATN codon. Cox1 begins with TTG, which may be a common codon in Zygentoma insects. Stem-loop structure can be observed in the largest non-coding region thought to be involved in the regulation of replication and transcription of the mitochondrial genome.
The Zygentoma are a small order of wingless hexapods; they are considered the sister-taxon of the winged insects (Pterygota) (Comandi et al. Citation2009). Currently, Zygentoma is divided into five recognized families (Cameron Citation2014), including Lepismatidae, Nicoletiidae, and Lepidotrichidae. The genus Ctenolepisma is the most diverse within the family Lepismatidae (Kahrarian et al. Citation2016). Ctenolepisma villosa is a species of family Lepismatidae, closely related to the silverfish and firebrats.
In this study, specimens of C. villosa were collected from Wannan Medical College on 1 December 2016 and preserved in alcohol. Ctenolepisma villosa were ground and total mtDNA was extracted from them using phenol-chloroform (Zhang and Alvarado Citation2018). Sequencing libraries were prepared by Shanghai Majorbio Bio-pharm Biotechnology Company (China) and sequenced with Illumina Hiseq sequencer (Minoche et al. Citation2011). Using Mega software (Tamura et al. Citation2007), 13 protein-coding genes were identified. We then manually mapped the cloverleaf secondary structure of 22 tRNA by comparing the anticodon and the secondary structures of the tRNAs of the other zygentoman species. Non-coding regions and two rRNA were confirmed using MAFFT software and a Mflod Web Page (Katoh and Standley Citation2013). Finally, we used Sequin software to annotate 37 genes and uploaded mitochondrial genome sequences online using Bankit (Bhatt et al. Citation2018).
The complete mitochondrial genome (mitogenome) of C. villosa (GenBank accession numbers: MK301436) was found to be composed of 15.488 bp and contained 37.2% A, 30.7% T, 20.6% C, 11.4% G, with higher overall A + T content (67.9%) observed for Zygentoma. The genome organization conforms with the putative ancestral insect gene arrangement (Clary and Wolstenholme Citation1985; Shao and Barker Citation2003).
13 PCGs were 11,195 bp in length. All protein-coding sequences with the exception of cox1 began with the typical ATN codon (seven with ATG, three with ATT, and two with ATA), and cox1 starts with TTG. Genes with TTG as their starting codon can also be found in the other two Zygentoma mitogenomes (Nardi Citation2003; Cook et al. Citation2005). It is speculated that TTG may be a common starting codon for certain genes within Zygentoma.
All the 22 tRNAs can be folded into an almost perfect cloverleaf secondary structure. Two rRNA genes are located on the N-strand and separated by the trnV gene. The rrnL is 1327 bp in length, and the rrnS is 800 bp long.
The largest non-coding region of C. villosa is 394 bp long. This falls well within the range of the control region of the other two Zygentoma: 397 bp in T. gertschi; 440 bp in A. formicaria (Nardi Citation2003; Cook et al. Citation2005; Comandi et al. Citation2009). In this region, the AT content is 72.1%, and a stem-loop structure was observed. A similar structure was also found in the other three Zygentoma species. For this reason, it can be speculated that this is the control region. This control region is thought to be involved in the regulation of replication, transcription, or both of the mitochondrial genome (Shadel and Clayton Citation1997).
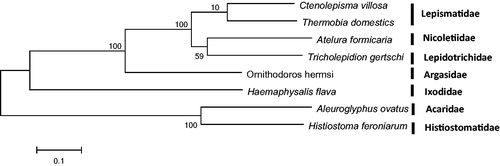
In this study, a maximum-likelihood phylogeny was inferred and showed the evolutionary relationship between Zygentoma species ().
Figure 1. Phylogram based on the mitogenome sequences of Ctenolepisma villosa (MK301436; this study) and three other Zygentoma species (plus four outgroup from Acari). The following mitochondrial genomes were used (accession numbers are in parentheses): Thermobia domestics (NC_006080), Atelura formicaria (EU084035), Tricholepidion gertschi (NC_005437), Ornithodoros hermsi (NC_039832), Haemaphysalis flava (MG604958), Aleuroglyphus ovatus (NC_023778), Histiostoma feroniarum (NC_038207).
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bhatt VD, Patel M, Joshi CG. 2018. Current trends in Bioinformatics: An Insight. Singapore(SG): Springer. Chapter 1, an insight of biological databases used in bioinformatics; p. 3–25.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 22:252–271.
- Comandi S, Carapelli A, Podsiadlowski L, Nardi F, Frati F. 2009. The complete mitochondrial genome of Atelura formicaria (Hexapoda: Zygentoma) and the phylogenetic relationships of basal insects. Gene. 439:25–34.
- Cook CE, Yue Q, Akam M. 2005. Mitochondrial genomes suggest that hexapods and crustaceans are mutually paraphyletic. Proc Biol Sci. 272:1295–1304.
- Kahrarian M, Molero R, Gaju M. 2016. The genus Ctenolepisma (Zygentoma: Lepismatidae) in Western Iran, with description of three new species. Zootaxa. 4093:217–230.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Minoche AE, Dohm JC, Himmelbauer H. 2011. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and Genome Analyzer systems. Genome Biol. 12:R112.
- Nardi F, Spinsanti G, Boore JL, Carapelli A, Dallai R, Frati F. 2003. Hexapod origins: monophyletic or paraphyletic?. Science. 299:1887–1889.
- Shadel GS, Clayton DA. 1997. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 66:409–435.
- Shao R, Barker SC. 2003. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol Biol Evol. 20:362–370.
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol Evol. 24:1596–1599.
- Zhang S, Alvarado AS. 2018. Planarian high molecular weight DNA isolation by spooling. Methods Mol Biol. 1774:277–284.
