Abstract
Here, we determined the complete mitochondrial genome of a bagrid catfish, Mystus rhegma. The whole mitogenome of M. rhegma was a circular molecule with 16,525 nucleotides, composed of 22 tRNA, 2 rRNA, 13 protein-coding genes, and a non-coding control region. The contents of A, T, C, and G are 30.89%, 26.00%, 27.43%, and 15.68%, respectively. In terms of AT and CG contents, the content of AT is obviously higher than that of CG, 56.89% and 43.11%, respectively. The phylogenetic analysis reveals that M. rhegma has a close relationship with M. mysticetus.
Keywords:
The Asian bagrid genus Mystus (Teleostei, Siluriformes) currently consists of 48 valid species and is widely distributed in tropical Asia (Froese and Pauly, Citation2019). At present, only two Mystus species (M. cavasius and M. vittatus) are reported for the complete mitochondrial genome data. Here, we sequenced the complete mitochondrial genome of another Mystus species, Mystus rhegma, which is a demersal freshwater catfish distributed in Cambodia, Laos, and Thailand (Froese and Pauly, Citation2019).
Samples were collected from Siem Reap in Cambodia (13.240391 N, 103.823090 E) and the specimens were deposited in the Southwest University Museum of Freshwater Fishes. The method described in Zeng et al. (Citation2012) was used to obtain the complete mitochondrial genome of M. rhegma. According to available gene data for Mystus species, we selected the mitochondrial cytochrome b gene sequences of 26 Mystus catfishes for subsequent analysis. The sequences were aligned and analyzed using MEGA7 (Kumar et al. Citation2016). The phylogenetic analysis based on the Bayesian inference (BI) method was conducted using MrBayes 3.2.6 (Ronquist et al. Citation2012).
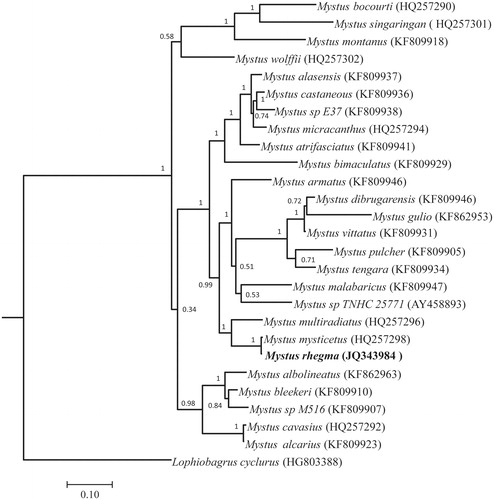
Figure 1. Bayesian inference phylogenetic tree of Mystus rhegma and the other 25 Mystus species using Lophiobagrus cyclurus as an outgroup. The posterior probabilities are labelled at each node.
The whole mitogenome of M. rhegma was a circular molecule with 16,525 nucleotides (GenBank accession No. JQ343984), composed of 22 tRNA, 2 rRNA (12S ribosomal RNA and 16S ribosomal RNA), 13 protein-coding genes (ND1, ND2, COX1, COX2, ATP8, ATP6, COX3, ND3, ND4L, ND4, ND5, ND6, and CYTB), and a non-coding control region. The majority of these genes are encoded on the heavy strand, apart from tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, tRNAPro, and ND6. The initiation codons of the 13 protein-coding genes are mainly ATG, while the COX1 gene starts with GTG. Moreover, most of these protein-coding genes have a common stop codon TAA. The contents of A, T, C, and G are 30.89% (5104), 26.00% (4297), 27.43% (4533), and 15.68% (2591), respectively. In terms of AT and CG contents, the content of AT is obviously higher than that of CG, 56.89% versus 43.11%.
The phylogenetic analysis reveals that M. rhegma has a close relationship with Mystus mysticetus, and these two species form a monophyletic group, which is closely related to Mystus multiradiatus ().
Acknowledgements
We thank Ms Dalin Ly (Royal University of Agriculture, Cambodia) for help in field sampling. We are grateful to Mr Qing Zeng for the laboratory work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Froese R, Pauly D, editors. 2019. FishBase. version (02/2019). http://www.fishbase.org.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Zeng Q, Ye H, Peng Z, Wang Z. 2012. Mitochondrial genome of Hemibagrus macropterus (Teleostei, Siluriformes). Mitochondrial DNA. 23:355–357.
