Abstract
Aridisia crenata Sims is one of the most popular ornamental plants in China, which leads to its high commercial values. Here, we report the complete chloroplast genome of A. crenata. The cp genome is 156,876 bp, including large single-copy (LSC) region of 86,421 bp, small single-copy (SSC) region of 18,347 bp, and two separated inverted regions of 26,054 bp, respectively. It contains 134 genes, including 87 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. The overall GC content of the cp genome is 37.1%. The phylogenetic analysis shows that A. crenata is sister to its congeneric species A. polysticta.
Ardisia crenata Sims is a welcome ornamental plant used in the south China because of its numerous light red fruits, symbolizing happiness, harvest, and fortune. It is the most widely distributed species in the genus Ardisia Swartz, found in most regions in the south of Yangtze River, as well as in Myanmar, Korea, Japan, India, Malaysia, Indonesia, Philippines, Cambodia, Vietnam, and Thailand. In this study, we reported the first chloroplast genome of A. crenata to better understand its genetic diversity and phylogenetic position in the future.
The fresh leaves of A. crenata were collected from Nanling National Forest Park in Shaoguan city, Guangdong Province. The specimen (voucher no.:Yxk160038) was deposited at the herbarium of South China Botanical Garden. Total genomic DNA was extracted from silica-gel dried leaves using a modified CTAB method (Doyle and Doyle Citation1987). The paired-end (2 × 150 bp) library was sequenced by Illumina Hiseq X Ten platform at Beijing Genomics Institute (Shenzhen, China). Totally 1.97 Gb clean data were obtained after removing low-quality reads and adaptor sequences. The complete chloroplast genome of A. crenata was assembled via a5-miseq (Coil et al. Citation2015), with manual adjustment and annotation using Geneious 11.0.3 (http://www.geneious.com, Kearse et al. Citation2012). The tRNA genes were annotated on ARAGORN (Laslett and Canback Citation2004). Phylogenetic analysis was performed using the maximum likelihood (ML) method. The aligned matrix of eight whole cp genomes (seven species in Primulaceae and one outgroup in Ebenaceae) was implemented in MAFFT (Katoh and Standley Citation2013). The maximum likelihood tree was constructed using RAxML (Stamatakis Citation2006) with 1000 bootstrap replicates.
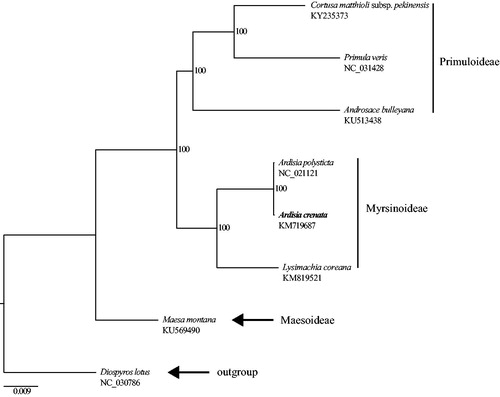
The A. crenata cp genome (GenBank accession no.: KM719687) is 156,876 bp in length. It has the typical quadripartite structure, including large single-copy (LSC) region of 86,421 bp, small single-copy (SSC) region of 18,347 bp, and two separated inverted regions (IRs) of 26,054 bp. 134 genes are identified in the whole cp genome, consisting of 87 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. The 63 protein-coding genes and 22 tRNA genes are located in LSC region (including one interregional gene rps19), while 11 protein-coding genes and 1 tRNA gene occur in the SSC region. All these eight rRNA genes are duplicated in the IR regions. IR regions contain six protein-coding genes and seven tRNA genes as well.19 genes contain one intron, while three genes (ycf3, clpP, and rps12) possess two introns. The first and other two parts of rps12 gene are located in the LSC region and IR regions, respectively. The overall GC content of the cp genome is 37.1%. The monophyly of Myrsinoideae is strongly supported (). In Myrsinoideae, A. crenata is sister to A. polysticta, and further clustered with Lysimachia coreana Nakai.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31:587–589.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Laslett D, Canback B. 2004. ARAGORN, a program for the detection of transfer RNA and transfer-messenger RNA genes in nucleotide sequences. Nucleic Acids Res. 32:11–16.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.