Abstract
We report on the structure and composition of the first complete mitochondrial genome of Stichaeus nozawae (typical prickleback fish (Zoarcales, Stichaeidae)), obtained by dideoxy (Sanger) sequencing technique. Genome has a classic organization with 13 genes, 2 rRNAs, control region, and 22 tRNAs, including additional tRNAs with atypical codons previously found in many eelpouts. Phylogenetic analysis confirms the species identity of S. nozawae and validate its recent divergence from S. grigorjewi with unusually low interspecific genetic distance.
Fishes of the genus Stichaeus are known to maintain the type characters of the pricklebacks (Zoarcales: Stichaeidae) which taxonomy is the issue of concern (Mecklenburg and Sheiko Citation2004; Pitruk et al. Citation2011; Parin et al. Citation2014; Radchenko Citation2015; Moreva et al. Citation2016). Two of the species, S. grigorjewi (SG) and S. nozawae (SN), distributed along the Sea of Japan and southern part of the Sea of Okhotsk (Mecklenburg and Sheiko Citation2004; Parin et al. Citation2014) have sharp discordance between morphological and molecular genetic species attributes. While SG and SN notably differ in external morphology and ecological features (Kolpakov and Klimkin Citation2004; Pitruk et al. Citation2011), their molecular genetic data show a little interspecific divergence which is comparable with the intraspecific variation of eelpouts and other teleost fishes (Ward Citation2009; Kwun and Kim Citation2013; Moreva et al. Citation2016; Turanov et al. Citation2016). This may indicate recent splitting of two lineages, where incipient species have not accumulated enough nucleotide substitutions to delineate (e.g. Shedko Citation2017). On the other hand, the mitochondrial control region (CR) sequences of eelpouts demonstrate the lack of saturation compared to protein-coding genes (Turanov, Lee, et al. Citation2019) and, therefore, it might express the natural patterns of evolutionary constrains of mitochondrial genome (mitogenome). To approach this problem from the genomic point of view, we obtained the first sequence of mitogenome for SN and compared with the mitogenome of SG (Turanov, Rutenko, et al. Citation2019).
Total DNA was isolated from the white muscle tissue of the specimen (MIMB 36617, female) collected in Vostok Bay of the Sea of Japan (42.89° N, 132.73° E) by gill nets in 2 May 2017. The mitogenome was obtained by dideoxy (Sanger) sequencing technique based on the set of newly designed primers with amplification of 27 overlapping regions. Primers were designed by Sliding Window-Based PSO Algorithm (Yang et al. Citation2011) on the basis of the mitogenome sequences available in GenBank for eelpouts. The resulted chromatograms were assembled into mitochondrial genome fragments using Geneious Trial (Kearse et al. Citation2012). The produced consensus sequence of complete genome was annotated using MitoAnnotator (Iwasaki et al. Citation2013) and MITOS Web Server (Bernt et al. Citation2013). To analyze the genetic polymorphism and divergence between S. grigorjewi and S. nozawae sequences, we used DnaSP v5 (Librado and Rozas Citation2009) and MEGA 7.0 (Kumar et al. Citation2016). The details of molecular phylogenetic analysis implemented to confirm the taxonomic identity and relative position of the obtained genome can be found in the previous mito communication (Turanov, Rutenko, et al. Citation2019).
The mitogenome of S. nozawae (MK561854) is 16,533 bp long, with following nucleotide base composition: T (27.5%), C (28.1%), A (26.6%), G (17.8%). Genome annotation revealed 13 coding fragments with most of the genes residing on the heavy strand while ND6 as well as eight tRNAs were encoded on the light-strand. It contained 22 tRNAs along with additional tRNA-Leu and tRNA-Ser, both having atypical codons as it was also found in many eelpouts (see Ayala et al. Citation2017; Rutenko et al. Citation2019). The annotation also revealed 16S and 12S rRNAs and CR, 862 pb in length.
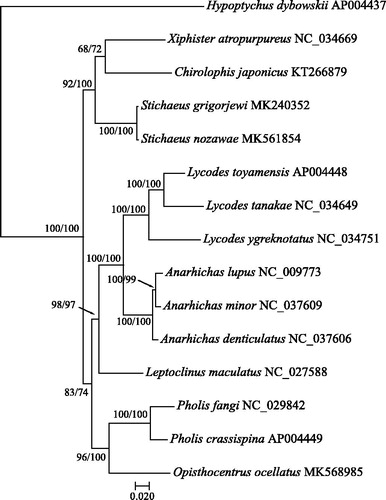
Uncorrected genetic distance between SG and SN was 0.4%, while between species of Anarhichas ranged from 1.6 to 2%, interspecific distances among Lycodes were 7.2–11%, and species divergence within Pholis was 5.2%. There were 71 variable sites observed and 3 indels (residing in CR) between SG and SN. Most of the nucleotide substitutions between them were distributed along ND1 (13), ND4 (9), Cyt-b (8), ND2 (8), ATPase 6 (5), ND6 (6) and COI (4). Molecular phylogeny () on the basis of 14 eelpouts species together with H. dybowskii (outgroup) demonstrates the closest position of SG and SN and the topological consistence with previous results (Rutenko et al. Citation2019; Turanov, Rutenko, et al. Citation2019). Thus, mitochondrial genomic data confirm the recent speciation between SG and SN and stimulate further research on the mechanisms of their origin.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ayala L, Becerra J, Boo GH, Calderon D, DiMarco A, Garcia A, Gonzales R, Hughey JR, Jimenez GA. 2017. The complete mitochondrial genome of Xiphister atropurpureus (Perciformes: Stichaeidae). Mitochondrial DNA Part B. 2:161–162.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, Nishida M. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kolpakov NV, Klimkin AF. 2004. Characteristics of the biology of the Grigoryev’s Stichaeus grigorjewi and Nozava’s S. nozawae (Stichaeidae) Stichaeus from the Northern Primorye. J Ichthyol. 44:637–644.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Kwun HJ, Kim JK. 2013. Molecular phylogeny and new classification of the genera Eulophias and Zoarchias (PISCES, Zoarcoidei). Mol Phylogenet Evol. 69:787–795.
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452.
- Mecklenburg CW, Sheiko BA. 2004. Family Stichaeidae Gill 1864 – Pricklebacks. Calif Acad Sci Annnotated Checklists Fishes. 35:1–36.
- Moreva IN, Radchenko OA, Neznanova SY, Petrovskaya AV, Borisenko SA. 2016. The relationships of Stichaeus nozawae (Jordan et Snyder, 1902) and Stichaeus grigorievi (Herzenstein, 1890) (Pisces: Stichaeidae) inferred from the data of genetic and karyological analyses and ultrastructural study of spermatozoa. Russ J Mar Biol. 42:471–480.
- Parin NV, Evseenko SA, Vasil’eva ED. 2014. Fishes of the Russian seas: annotated catalogue. Moscow: KMK Scientific Press.
- Pitruk DL, Lavrova TV, Zemnukhov VV. 2011. A morphological description of the brown shanny Stichaeus fuscus Miki et Maruyama, 1986 (Perciformes: Stichaeidae). Russ J Mar Biol. 37:458–463.
- Radchenko OA. 2015. The system of the suborder Zoarcoidei (Pisces, Perciformes) as inferred from molecular genetic data. Russ J Genet. 51:1096–1112.
- Rutenko OA, Turanov SV, Kartavtsev YP. 2019. Complete mitochondrial genome of ocellated blenny, Opisthocentrus ocellatus (Tilesius, 1811) (Zoarcales: Opisthocentidae). Mitochondrial DNA Part B. 4:1553–1555.
- Shedko SV. 2017. The low level of differences between mitogenomes of the Sakhalin sturgeon Acipenser mikadoi Hilgendorf, 1892 and the green sturgeon A. medirostris Ayeres, 1854 (Acipenseridae) indicates their recent divergence. Russ J Mar Biol. 43:176–179.
- Turanov SV, Kartavtsev YP, Lipinsky VV, Zemnukhov VV, Balanov AA, Lee Y-H, Jeong D. 2016. DNA-barcoding of perch-like fishes (Actinopterygii: Perciformes) from far-eastern seas of Russia with taxonomic remarks for some groups. Mitochondrial DNA. 2:1188–1209.
- Turanov SV, Lee Y-H, Kartavtsev YP. 2019. Structure, evolution and phylogenetic informativeness of eelpouts (Cottoidei: Zoarcales) mitochondrial control region sequences. Mitochondrial DNA Part A. 30:264–272.
- Turanov SV, Rutenko OA, Kartavtsev YP. 2019. Complete mitochondrial genome of Stichaeus grigorjewi Herzenstein, 1890 (Zoarcales: Stichaeidae). Mitochondrial DNA Part B. 4:899–901.
- Ward RD. 2009. DNA barcode divergence among species and genera of birds and fishes. Mol Ecol Resour. 9:1077–1085.
- Yang C-H, Chang H-W, Ho C-H, Chou Y-C, Chuang L-Y. 2011. Conserved PCR primer set designing for closely-related species to complete mitochondrial genome sequencing using a sliding window-based PSO algorithm. PLoS One. 6:e17729.