Abstract
Senecio vulgaris is an annual or biennial invasive weed. The complete chloroplast is 150,806 bp in length, containing a large single-copy region of 82,950 bp and a small single-copy region of 18,214 bp. It encodes a set of 113 genes, consisting of 79 protein-coding, 30 tRNA, and 4 rRNA genes. 14 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rps12, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC) have a single intron, and two genes (clpP and ycf3) possess double introns. Phylogenetic analysis confirmed that the cp genome of S. vulgaris is closely related to that of Jacobaea vulgaris.
Senecio vulgaris L. (Asteraceae), an annual or biennial invasive weed most probably originated from southern Europe, is a small herb with a strong expansion ability for its short life cycle (8 weeks), efficient seed germination ability and high reproductive ability (Robinson et al. Citation2003; Figueroa et al. Citation2007). This species is introduced into northeast China in the nineteenth century and is now widely distributed in open hillsides, grasslands, farmlands, and roadsides at altitudes of 300–2300 m in China (Robinson et al. Citation2003). This invasive species may displace the growth and development of local species, destroy the structure and distribution of native plant communities, and cause biodiversity loss, health damage and economic harm (Blackburn et al. Citation2011). It is listed in the Checklist of the Invasive Plants in China (Cheng et al. Citation2017; Zhu et al. Citation2017). Senecio vulgaris also has important medicinal and ecological value (Wei et al. Citation2003; Xiong et al. Citation2014). Some studies confirm that it is a new medicinal plant (Xiong et al. Citation2014) and also a Cd super-enriched plant (Wei et al. Citation2003). In this study, we characterized the complete chloroplast genome sequence of S. vulgaris as a resource for future genetic studies on this and other related species, which may provide valuable guidance for the management and utilization of S. vulgaris.
Fresh leaves of S. vulgaris were obtained from the Kangding County, Ganzi Tibetan Autonomous Prefecture, Sichuan Province of China (101°29′33″E, 30°2′36″N) with voucher specimen also deposited at the Anqing Normal University Herbarium (AQ2018082206). After DNA extraction, high-throughput DNA sequencing (pair-end 150 bp) was conducted on an Illumina Hiseq X Ten platform (Illumina, CA, USA). Approximately 5.2 Gb of sequence data were generated and used for the assembly of cp genome with MITObim v1.9 (Hahn et al. Citation2013). The cp genome of Jacobaea vulgaris (HQ234669) (Leonie et al. Citation2011) was included as the initial reference.
The complete chloroplast genome sequence of S. vulgaris (GenBank accession MK 654722) is 150,806 base pairs (bp) in length, containing a large single-copy (LSC) region of 82,950 bp, a small single-copy (SSC) region of 18,214 bp, which are separated by two inverted repeat (IR) regions of 24,821 bp. The overall GC-content of the whole plastome is 37.30%. A total of 113 genes are predicted, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Among all of these genes, 2 genes (clpP and ycf3) possess double introns, and 14 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rps12, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC) have a single intron.
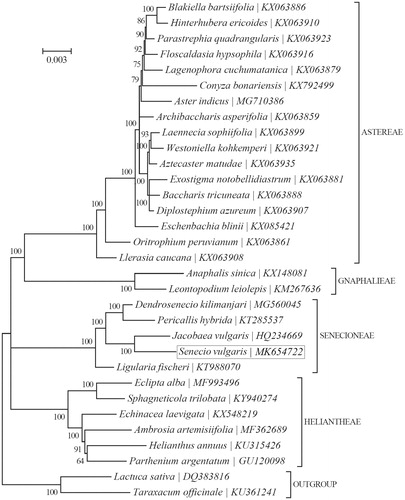
Phylogeny of 30 species within the subfamily Asteroideae based on the Bayesian inference (BI) of the concatenated chloroplast protein-coding sequences were studied. The GTR + G + I model was employed as the best-fit nucleotide substitution model as suggested by TOPALi v2.5. Phylogenetic analysis showed that S. vulgaris clustered together with Jacobaea vulgaris (). The chloroplast resource may provide valuable guidelines for the management and utilization of S. vulgaris.
Figure 1. Phylogeny of 30 species within the subfamily Asteroideae based on the Bayesian inference (BI) of the concatenated chloroplast protein-coding sequences. The GTR + G + I model was employed as the best-fit nucleotide substitution model as suggested by TOPALi v2.5. The support values are placed next to the branches. The tribe-level taxonomy is also shown.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Blackburn TM, Pyšek P, Bacher S, Carlton JT, Duncan RP, Jarošík V, Wilson JRU, Richardson DM. 2011. A proposed unified framework for biological invasions. Trends Ecol Evol. 26:333–339.
- Cheng D, Nguyen VT, Ndihokubwayo N, Ge J, Mulder P. 2017. Pyrrolizidine alkaloid variation in Senecio vulgaris populations from native and invasive ranges. PeerJ. 5:e3686.
- Figueroa R, Doohan D, Cardina J, Harrison K. 2007. Common groundsel (Senecio vulgaris) seed longevity and seedling emergence. Weed Sci. 55:187–192.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Leonie D, Barbara G, Youri L, Yavuz A, Thomas CAW, Klaas V. 2011. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 18:93–105.
- Robinson D, O’Donovan J, Sharma M, Doohan D, Figueroa R. 2003. The biology of Canadian weeds. 123. Senecio vulgaris L. Can J Plant Sci. 83:629–644.
- Wei S, Zhou Q, Wang X. 2003. Characteristics of 18 species of weed hyperaccumulating heavy metals in contaminated soils. J. Basic Sci Eng. 11:152–160. [in Chinese with English abstract].
- Xiong AZ, Fang LX, Yang X, Yang F, Qi M, Kang H, Yang L, Tsim KW, Wang ZT. 2014. An application of target profiling analyses in the hepatotoxicity assessment of herbal medicines: comparative characteristic fingerprint and bile acid profiling of Senecio vulgaris L.and Senecio scandens Buch.Ham. Anal Bioanal Chem. 406:7715–7727.
- Zhu BR, Barrett SCH, Zhang DY, Liao WJ. 2017. Invasion genetics of Senecio vulgaris: loss of genetic diversity characterizes the invasion of a selfing annual, despite multiple introductions. Biol Invasions. 19:255–267.
