Abstract
Stellera chamaejasme is a toxic perennial herb found in open and dry places in East Asia and has the potential for new drugs for anti-inflammatory, analgesic, and cutaneous wound-healing purposes. Here, we present the complete chloroplast genome of S. chamaejasme. Its length is 173,381 bp and has four subregions; 86,769 bp of large-single-copy and 2,858 bp of small-single-copy regions are separated by 41,877 bp of inverted repeat regions, including 135 genes (89 protein-coding genes, 8 rRNAs, and 38 tRNAs). Phylogenetic trees show that S. chamaejasme is a sister to Daphne kiusiana, supporting morphological affinity of the two species.
Stellera chamaejasme L. (Thymelaeaceae) is a toxic perennial herb found in open and dry places such as grasslands and meadows in Bhutan, Nepal, China, Mongolia, the Russian Far East, and Korea (Wang and Gilbert Citation2007; Zhang et al. Citation2015). It can easily be recognized by having caespitose and unbranched stems; a terminal head with an involucre, yellow, pink, or red apetalous flowers; and an articulate hypanthium within Thymelaeaceae. Stellera chamaejasme has been widely used in traditional Chinese medicine for tumors, tuberculosis, and psoriasis (Asada et al. Citation2011; Zhao et al. Citation2013). Extracts from S. chamaejasme have anti-inflammatory, analgesic, and cutaneous wound-healing activities (Kim et al. Citation2017). Stellera chamaejasme is locally common throughout its range, except in Korea, where populations are only found in the eastern part of Jejudo Island. Plants of S. chamaejasme in this region, however, were overexploited for ornamental purposes, and it was recently designated as an endangered species in Korea (http://species.nibr.go.kr).
The complete chloroplast genome of S. chamaejasme was also determined to be useful during the development of new drugs and in conservation biology associated with this species. Total DNA of S. chamaejasme isolated from Jejudo Island, Korea (Voucher in the Herbarium of the National Institute of Biological Resources, Korea (KB); Oh 7533) was extracted from fresh leaves using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). The genome was sequenced using HiSeq4000 at Macrogen Inc., Korea, and de novo assembly and confirmation were performed by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAP GapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for annotation based on the Daphne kiusiana chloroplast genome (NC035896; Cho et al. Citation2018).
The chloroplast genome of S. chamaejasme (GenBank accession is MK681211) is 173,381 bp (the GC ratio is 36.7%) and has four subregions; 86,769 bp of large-single-copy (LSC; 34.8%) and 2858 bp of small-single-copy (SSC; 29.3%) regions are separated by 41,877 bp each of inverted repeats (IR; 38.8%). It contains 135 genes (89 protein-coding genes, 8 rRNAs, and 38 tRNAs), with 28 genes (16 protein-coding genes, 4 rRNAs, and 8 tRNAs) duplicated in the IR regions. The length of SSC of S. chamaejasme is extremely short. The lengths of SSC of D. kiusiana, Pimelea aquilonia, Aquilaria yunnanensis, and A. malaccensis are also short (around 3,000 bp), suggesting that this structural characteristic may be common in the family. The IR of S. chamaejasme is very long. The expansion of IR is associated with the shortening of SSC (Zhu et al. Citation2016).
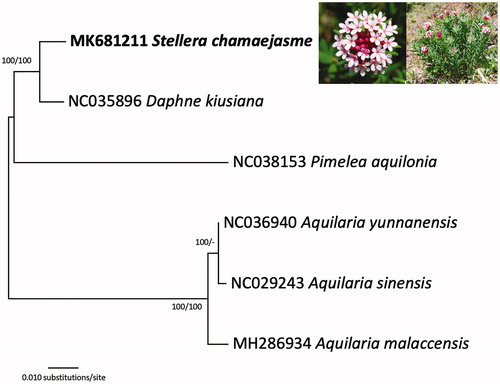
Six Thymelaeaceae chloroplast genomes were used for constructing Neighbor-joining (with 10,000 bootstrap repeats) and Maximum-likelihood (with 1,000 bootstrap repeats) phylogenic trees using MEGA X (Kumar et al. Citation2018) after aligning whole chloroplast genomes using MAFFT 7.388 (Katoh and Standley Citation2013). The phylogenetic trees show that S. chamaejasme is a sister to D. kiusiana (), supporting the morphological affinity of the two species (Lee and Oh Citation2017).
Figure 1. A Neighbor-joining (with 10,000 bootstrap replicates) phylogenetic tree of six Thymelaeaceae chloroplast genomes, specifically Stellera chamaejasme (MK681211 in this study), Daphne kiusiana (NC035896), Pimelea aquilonia (NC038153), Aquilaria yunnanensis (NC036940), Aquilaria malaccensis (MH2866934), and Aquilaria sinensis (NC029243). The phylogenetic tree based on the Maximum-likelihood method (with 1,000 bootstrap replicates) was identical to the NJ tree. The numbers above the branches indicate the corresponding bootstrap support values from the Maximum-likelihood and Neighbor-joining methods.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Asada Y, Sukemori A, Watanabe T, Malla KJ, Yoshikawa T, Li W, Koike K, Chen C-H, Akiyama T, Qian K, et al. 2011. Stelleralides A-C, novel potent anti-HIV daphnane-type diterpenoids from Stellera chamaejasme L. Org Lett. 13:2904–2907.
- Cho W-B, Han E-K, Choi G, Lee J-H. 2018. The complete chloroplast genome of Daphne kiusiana, an evergreen broad-leaved shrub on Jeju Island. Conserv Genet Res. 10:103–106.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim M, Lee HJ, Randy A, Yun JH, Oh S-R, Nho CW. 2017. Stellera chamaejasme and its constituents induce cutaneous wound healing and anti-inflammatory activities. Sci Rep. 7:42490.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Lee JJ, Oh S-H. 2017. A comparative morphological study of Thymelaeaceae in Korea. Korean J Pl Taxon. 47:207–221.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 1303.3997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Wang Y, Gilbert MG. 2007. Stellera Linnaeus. Flora of China, Vol. 13. Beijing, China: Science Press.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research. 18:821–829.
- Zhang Y-H, Zhang S-D, Ling L-Z. 2015. De novo transcriptome analysis to identify flavonoid biosynthesis genes in Stellera chamaejasme. Plant Gene. 4:64–68.
- Zhao M, Gao X, Wang J, He X, Han B. 2013. A review of the most economically important poisonous plants to the livestock industry on temperate grasslands of China. J Appl Toxicol. 33:9–17.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
- Zhu A, Guo W, Gupta S, Fan W, Mower JP. 2016. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 209:1747–1756.
