Abstract
In this study, the complete mitochondrial genome of Dermogenys pusilla was firstly determined. The complete genome is 16,529 bp in length. It contains 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, a putative control region, and 1 origin of replication on the light-strand. The gene organization, nucleotide composition, and codon usage are similar to other Hemiramphidae. The overall nucleotide composition was 30.73% A, 28.18% T, 26.48% C, and 14.61% G, respectively. All protein-coding genes started with Met, ND1, ND2, CO1, ATP8, ATP6, CO3, ND3, ND4L, ND5 and ND6 ended by TAA as a stop codon, ND4 ended by AGA as a stop codon, CO2 and Cytb use an incomplete stop codon T. The lengths of 12S ribosomal RNA and 16S ribosomal RNA are 941 bp and 1670 bp, respectively. The length of control region is 862 bp, ranging from 15,668 bp to 16,529 bp. The complete mitochondrial genome sequence provided here would help us understand the evolution of ratite and conservation genetics of D. pusilla in the future.
Dermogenys pusilla is a species belonging to the family Hemiramphidae, which lives in rivers, rivulets, canals, drains, ponds and lakes, rarely found in the sea (Meisner and Burns Citation1997; Bruyn et al. Citation2010). There were few reports about its basic biology data including genetic information. Considering its ecological importance and commercial value, to gain its molecular information, we described the complete mitogenome of D. pusilla, and explored the phylogenetic relationship within Hemiramphidae, provide a new thought for propagation and breeding of this species.
The specimen was collected from Pa Sak River, Thailand (15°19′01″N, 101°12′56″E) and stored in a refrigerator of –80 °C at National Engineering Research Center for Marine Aquaculture. Total genomic DNA was extracted from the muscle tissue of individual using the phenol–chloroform method (Barnett and Larson Citation2012). The calculation of base composition and phylogenetic construction was conducted by MEGA 6.0 software (Tamura et al. Citation2013). Similar to the typical mitogenome of vertebrates, the mitogenome of D. pusilla is a closed double-stranded circular molecule of 16,529 bp (GenBank accession No. MK034755), which contains 13 protein-coding genes, 2 ribosomal RNA genes, 22 tRNA genes, and 2 main non-coding regions (Boore Citation1999; Zhu, Gong, et al. Citation2018; Zhu, Lu, et al. Citation2018). The overall base composition is 30.73%, 28.18%, 26.48%, and 14.61% for A, T, C, and G, respectively. Most mitochondrial genes are encoded on the H-strand except for ND6 and eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu and tRNA-Pro), which are encoded on the L-strand. All of them use the initiation codon ATG except COI uses GTG, which is quite common in vertebrate mtDNA (Miya et al. Citation2001; Li et al. Citation2016). Most of them have TAA as the stop codon, whereas ND4 ends with AGA, and two protein-coding genes (COII and Cytb) ended with an incomplete stop codon T. The 12S rRNA and 16S rRNA are 941 and 1670 bp, which are both located in the typical positions between tRNA-Phe and tRNA-Leu(UAA), separated by tRNA-Val.
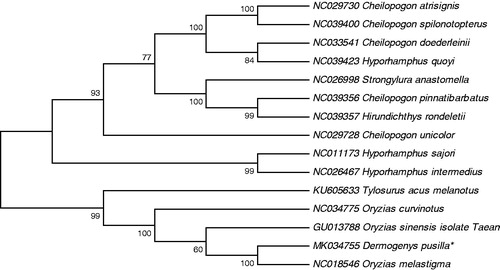
To explore the phylogenetic position of this D. pusilla, a phylogenetic tree was constructed based on the NJ analysis of 12 PCGs encoded by the heavy strand. The results of the present study suggest that D. pusilla has a closest relationship with Oryzias melatigma, highly supported by a bootstrap value of 100 (), which are consistent with the results based on morphology and other molecular methods.
Figure 1. Neighbour joining (NJ) tree of 15 Hemiramphidae species based on 12 PCGs. The bootstrap values are based on 10,000 resamplings. The number at each node is the bootstrap probability. The number before the species name is the GenBank accession number. The genome sequence in this study is labelled with an asterisk.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Barnett R, Larson G. 2012. A phenol-chloroform protocol for extracting DNA from ancient samples. Methods Mol Biol. 840:13–19.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Bruyn MD, Grail W, Barlow A, Carvalho GR. 2010. Anonymous nuclear markers for Southeast Asian halfbeak fishes (dermogenys). Conserv Genet Resour. 2:325–327.
- Li Y, Xue D, Gao T, Liu J. 2016. Genetic diversity and population structure of the rough skin sculpin (Trachidermus fasciatus Heckel) inferred from microsatellite analyses: implications for its conservation and management. Conserv Genet. 17:921–930.
- Meisner AD, Burns JR. 1997. Viviparity in the halfbeak genera Dermogenys and Nomorhamphus (Teleostei: Hemiramphidae). J Morphol. 234:295–317.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences . Mol Biol Evol. 18:1993–2009.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zhu K, Gong L, Jiang L, Liu L, Lü Z, Liu B-j. 2018. Phylogenetic analysis of the complete mitochondrial genome of Anguilla japonica (Anguilliformes, Anguillidae). Mitochondrial DNA Part B. 3:536–537.
- Zhu K, Lu Z, Liu L, Gong L, Liu B. 2018. The complete mitochondrial genome of Trachidermus fasciatus (Scorpaeniformes: Cottidae) and phylogenetic studies of Cottidae. Mitochondrial DNA Part B. 3:301–302.
