Abstract
Solanum demissum is a wild tuber-bearing species belonging to Solanaceae family. The complete chloroplast genome of S. demissum was constituted by de novo assembly using a small amount of whole genome sequencing data. The chloroplast genome of S. demissum was circular DNA molecule with a length of 155,558 bp and consisted of 85,999 bp of large single copy, 18,373 bp of small single copy, and 25,593 bp of a pair of inverted repeat regions. A total of 158 genes were annotated including 105 protein-coding genes, 45 tRNA genes, and 8 rRNA genes. Maximum likelihood phylogenetic analysis with 30 Solanaceae species revealed that S. demissum is most closely grouped with S. hougasii and S. stoloniferum.
The Mexican hexaploid species, Solanum demissum is one of the tuber-bearing wild relatives to the cultivated potato, S. tuberosum and the most famous resource for the late blight disease caused by Phytophthora infestans (Ross Citation1986; Plastied and Hoopes Citation1989). Its EBN (Endosperm Balanced Number) value of four theoretically makes it directly crossable for breeding purposes with cultivated tetraploid potatoes (Hawkes Citation1990; Ortiz and Ehlenfeldt Citation1992; Cho et al. Citation1997; Spooner et al. Citation2014; Haynes and Qu Citation2016), and the species can easily be used to obtain hybrids with S. tuberosum (Dionne Citation1961; Sanetomo et al. Citation2011). Moreover, it has been considered as an allohexaploid with three well-differentiated genomes (Ono et al. Citation2016) and its nuclear genome composition has evolutionally been identified by a gene and GISH analysis (Spooner et al. Citation2008; Pendinen et al. Citation2012). However, plastid genome of the species was rarely studied. Information of chloroplast genome of the species obtained in this study will provide an opportunity to investigate more detailed evolutionary and breeding aspects.
The accession S. demissum (PI218047) was originally collected in Mexico by International Potato Centre. The seeds of the species was provided by Highland Agriculture Research Institute and stored at Daegu University, South Korea. After seedling, a total genomic DNA was extracted and used to construct a paired-end (PE) genomic library according to the standard protocol (Illumina, San Diego, USA). Sequencing was conducted using a HiSeq2000 at Macrogen (http://www.macrogen.com/kor/). Low-quality bases with raw scores of 20 or less were removed and ∼2.5Gbp of high-quality of PE reads were obtained. The sequences were assembled by a CLC genome assembler (CLC Inc, Rarhus, Denmark) (Kim et al. Citation2015). The reference sequence of S. berthaultii (KY419708, Park Citation2017) was used to retrieve principal contigs representing the chloroplast genome from the total contigs using Nucmer (Kurtz et al. Citation2004). The representative chloroplast contigs were arranged in the order based on BLASTZ analysis (Schwartz et al. Citation2003) with the reference sequence and connected to a single draft sequence by joining the overlapping terminal sequences. The chloroplast genes were predicted using DOGMA (Wyman et al. Citation2004) and BLAST searches.
The complete chloroplast genome of S. demissum (MK036508) was 155,558 bp in length including 25,593 bp inverted repeats regions separated by small single-copy region of 18,373 bp and large single-copy region of 85,999 bp with the typical quadripartite structure of most plastids, and the structure and gene features were typically identical to those of higher plants. A total of 158 genes with an average size of 583.0 bp were annotated including 105 protein-coding genes with an average size of 764.6bp, 45 tRNA gene,s and 8 rRNA genes. The overall GC content was 37.25%.
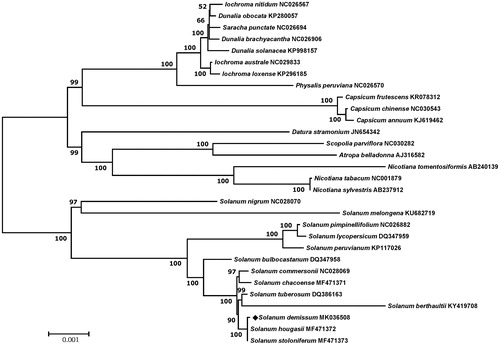
Chloroplast coding sequences of S. demissum and 29 published species in Solanaceae family were analyzed to generate a phylogenetic tree using a maximum likelihood method in MEGA 6.0 (Tamura et al. Citation2013). According to the phylogenetic tree, S. demissum belonged to the same clade in Solanum species and it was most closely grouped with S. hougasii and S. stoloniferum ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cho HM, Kim-Lee HY, Om YH, Kim JK. 1997. Influence of endosperm balance number (EBN) in interploidal and interspecific crosses between Solanum tuberosum dihaploids and wild species. Korean J Breed. 29:154–161.
- Dionne LA. 1961. Cytoplasmic sterility in derivatives of Solanum demissum. Am Potato J. 38:117–120.
- Hawkes JG. 1990. The potato: evolution, biodiversity and genetic resources. London, UK: Belhaven Press.
- Haynes KG, Qu X. 2016. Late blight and early blight resistance from Solanum hougasii introgressed into Solanum tuberosum. Am J Potato Res. 93:86–95.
- Kim K, Lee SC, Lee J, Lee H, Joh HJ, Kim NH, Park HS, Yang TJ. 2015. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 10:e0117159.
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12.
- Ono S, Sanetomo R, Hosaka K. 2016. Genetic transmission of Solanum demissum (2n = 6x = 72) chromosomes from a pentaploid hybrid of S. tuberosum (2n = 4x = 48) into aneuploid BC1 progeny. Euphytica. 207:149–168.
- Ortiz R, Ehlenfeldt MK. 1992. The importance of endorsperm balance number in potato breeding and the evolutino of tuber-bearing Solanum species. Euphytica. 60:105–113.
- Park T-H. 2017. The complete chloroplast genome sequence of Solanum berthaultii, one of the potato wild relative species. Mitochondrial DNA Part B. 2:88–89.
- Pendinen G, Spooner DM, Jiang J, Gavrilenko T. 2012. Genomic in situ hybridization reveals both auto- and allopolyploid origins of different North and Central American hexaploid potato (Solanum sect. Petota) species. Genome. 55:407–415.
- Plastied RL, Hoopes RW. 1989. The past record and future prospects for the use of exotic potato gerplasm. Am Potato J. 66:603–627.
- Ross H. 1986. Potato breeding: problems and perspectives. Berlin and Hamburg, Germany: Verlag Paul Parey.
- Sanetomo R, Ono S, Hosaka K. 2011. Characterization of crossability in the crosses between Solanum demissum and S. tuberosum, and the F1 and BC1 progenies. Am J Potato Res. 88:500–510.
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. 2003. Human-mouse alignments with BLASTZ. Genome Res. 13:103–107.
- Spooner DM, Ghislain M, Simon R, Jansky SH, Gavrilenko T. 2014. Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. Bot Rev. 80:283–383.
- Spooner DM, Rodríguez F, Polgár Z, Ballard HE, Jr, Jansky SH. 2008. Genomic origins of potato polyploids: GBSSI gene sequencing data. Crop Sci. 48:S27–S36.
- Tamura K, STecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.