Abstract
The complete mitochondrial DNA genome of Anabarilius brevianalis was determined. The entire mitochondrial genome is 16,610 bp in length and contains 13 protein-coding genes, 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and 1 control region (CR). The gene arrangement, gene numbers, base composition, and codon usage pattern of this species were similar to the typical Cultrinae fish mitochondrial genomes. Maximum likelihood phylogenetic analysis based on the complete mitochondrial sequence shows the A. brevianalis clustered with A. grahami are closely related to Hemiculter branch within the subfamily Cultrinae.
Anabarilius brevianalis Zhou and Cui belongs to the genus Anabarilius within the subfamily Cultrinae of the family Cyprinidae, which is an endemic species to the Canyu River, a left tributary of the upper Yangtze River of China (Zhou and Cui Citation1992). Because of over-exploitation and damage to spawning beds, wild A. brevianalis has not been sampled in the field in recent years except for some individuals by artificial reproduction and releasing from a small local culture base. It has been listed as a key protected species in Sichuan Province of China in 2000, as a vulnerable species in the Redlist of China’s Biodiversity by Ministry of Environmental Protection and Chinese Academy of Sciences in 2015 (www.mee.gov.cn) and as a critically endangered species in Zhang et al. (Citation2015). No other studies on this species were conducted in recent years, except for it was introduced as a new species in the upper Yangtze River in 1992 (Zhou and Cui Citation1992) and evaluated as vulnerable species (Zhang et al. Citation2015).
In this study, the complete mtDNA sequence of A. brevianalis was determined. The samples were collected from the Canyu River, a left tributary of the upper Yangtze River of China in 2018. One female and one male were used to determine the mitochondrial genome. After sampling, the specimens were stored in 100% ethanol and kept in the Yangtze River Fisheries Research Institute. The genomic DNA was isolated from fin tissue by a salt extraction protocol. Fifteen primers were used to amplify the segments. The protein-coding genes, rRNA genes, tRNA genes, and one control region of the mitochondrial genome were annotated using MitoAnnotator (Iwasaki et al. Citation2013). The complete mitochondrial genome sequence of A. brevianalis was 16,610 bp in length (GenBank Accession number MK757491), containing 13 protein-coding genes, 2 ribosomal RNA genes (rRNA), 22 transfer (tRNA) genes, and 1 control region. There were no differences in the complete mitochondrial genome sequence between female and male. The overall nucleotide composition is 28.3% A, 24.6% T, 28.4% C, and 18.7% G, exhibiting the A + T-rich content (52.9%). Most of the A. brevianalis mitochondrial genes were encoded on the heavy strand (H-strand) except for ND6 and eight tRNA genes (tRNA-Gln, -Ala, -Asn, -Cys, -Tyr, -Ser, -Glu, -Pro) which were encoded on the light strand (L-strand). The gene arrangement, gene numbers, base composition, and codon usage pattern of this species were similar to the typical Cultrinae fish mitochondrial genomes (Liu et al. Citation2016; Zou et al. Citation2019; Wang et al. Citation2018). Among 13 protein-coding genes, six mitochondrial genes (ND1, COI, ATP8, ND4L, ND5, and ND6) are encoded by the typical TAA or TAG stop codons, except for genes ND2, COII, ATP6, COIII, ND3, ND4 and Cyt b which are encoded by an incomplete stop codon T(AA). The control region is a 929 bp sequence located between the tRNA-Pro and tRNA-Phe genes.
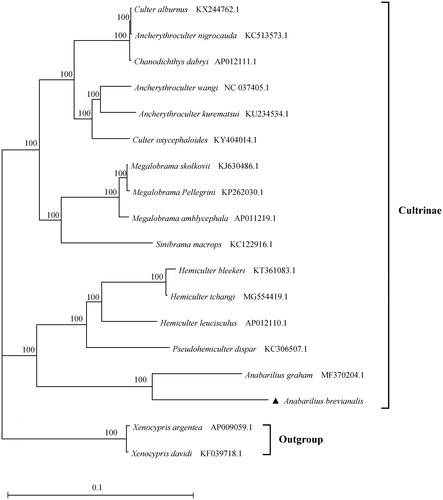
Limited mitochondrial genome sequences of the genus Anabarilius were reported, and the only reports available were for A. grahami (Wang et al. Citation2018) and A. Liui yalongensis (Zeng et al. Citation2019). However, the mitochondrial genome sequence of A. Liui yalongensis is unavailable in the NCBI website to date, hence was not included in the present phylogenetic analysis. In order to explore the evolutionary status of A. brevianalis within the subfamily Cultrinae, a maximum likelihood (ML) tree was constructed using PhyML 3.0 (Guindon et al. Citation2010). Xenocypris davidi and Xenocypris argentea were used as outgroups for tree rooting. Phylogenetic tree demonstrated that the A. brevianalis clustered with A. grahami are closely related to the Hemiculter branch within the subfamily Cultrinae (), which was in agreement with Wang et al. (Citation2018) and Zeng et al. (Citation2019). It was also in agreement with the phylogeny of subfamily Cultrinae given by Yue and Luo (Citation1996) based on key morphological traits. In conclusion, the complete mitochondrial genome of A. brevianalis reported in this study would facilitate further investigations of molecular evolution of species in the subfamily Cultrinae and in the genus Anabarilius, and also be beneficial for the management and conservation.
Acknowledgements
The authors are grateful to J. Zou and L. Wang for their help in the sampling of experimental fish.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, Nishida M. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Liu XQ, Xiao K, Zhao X, Liu JJ, Zhang X, Guo WT, Chen L, Du HJ. 2016. The complete mitochondrial genome of the Megalobrama Pellegrini (Teleostei: Cyprinidae). Mitochondrial DNA. 27:3069–3070.
- Wang Y, Wen H, Liu H, Xiong F. 2018. Complete mitochondrial genome and phylogenetic analysis of Anabarilius grahami (Teleostei, Cyprinidae, Cultrinae). Conserv Genet Resour. 10:301–303.
- Yue PQ, Luo Y. 1996. Preliminary studies on phylogeny of subfamily Cultrinae (Cypriniformes: Cyprinidae). Acta Hydrobiol Sin. 20:182–185.
- Zeng LW, Yang K, Wen AX, Xie M, Yao YF, Xu HL, Zhu GX, Wang Q, Jiang YZ, He T, Wu JY. 2019. Complete mitochondrial genome of the endangered fish Anabarilius liui yalongensis (Teleostei, Cyprinidae, Cultrinae). Mitochondrial DNA Part B. 4:21–22.
- Zhang X, Gao X, Wang JW, Cao WX. 2015. Extinction risk and conservation priority analyses for 64 endemic fishes in the upper Yangtze River, China. Environ Biol Fish. 98:261–272.
- Zhou W, Cui GH. 1992. Anabarilius brevianalis, a new species from the Jinshajiang River basin, China (Teleostei: Cyprinidae. ). Ichthyol Explor Fres. 3:49–54.
- Zou YC, Liu T, Li Q, Wen ZY, Qin CJ, Li R, Wang DS. 2019. Complete mitochondrial genome of Hemiculter tchangi (Cypriniformes, Cyprinidae). Conserv Genet Resour. 11:1–4.