Abstract
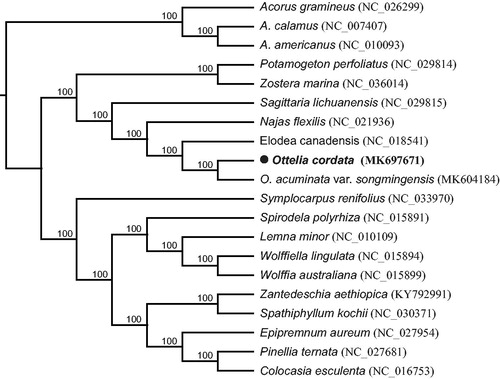
Ottelia cordata (Wallich) Dandy (Hydrocharitaceae) is an endangered aquatic macrophyte. The complete chloroplast genome sequence of O. cordata was generated by de novo assembly using whole genome next-generation sequencing. The complete chloroplast genome was 157,879 bp in length and constructed out of four parts – a large single-copy (LSC) region of 87,672 bp, a small single-copy (SSC) region of 20,265 bp, and two inverted repeat (IRa and IRb) regions of 24,971 bp each. The genome annotation predicted a total of 117 genes, including 72 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis of 20 selected chloroplast genomes revealed that sampled species of Hydrocharitaceae formed a monophyletic clade, in which O. cordata and O. acuminata var. songmingensis formed a sister clade to Elodea canadensis.
Ottelia cordata (Wallich) Dandy, belonging to the family Hydrocharitaceae, is an endangered aquatic macrophyte distributed in China, Cambodia, Myanmar, and Thailand (Wang et al. Citation2010). This species is dioecious, insect-pollinated, and shows leaves dimorphism in submerged and floating leaves. In China, O. cordata is found only in Haikou, Hainan province, with very narrow distribution range (Yu et al. Citation1998). In recent years, many populations have perished and the species range has been dramatically reduced due to multiple factors, such as water eutrophication, wetland destruction. Ottelia cordata has been categorized as an endangered species in the List of Key Protected Wild Plants in China. Several wetland reserves have been established in Hainan Province, such as the Nayang Wetland Reserve and Changwangxi Wetland Reserve, to protect this species. In this study, we reported the complete cp genome of O. cordata to provide genomic resource for further conservation genetics and phylogenetic analysis.
The fresh leaves of O. cordata were collected from Nayang Wetland Reserve, Haikou (19°56′34″N, 110°23′51″E), and voucher specimens (ZhangHN14) were deposited in the Herbarium of Yunnan Normal University. A sequence library was constructed and sequencing was performed using the Illumina HiSeq 2500-PE150 platform (Illumina, CA, USA). All raw reads were filtered using NGS QC Toolkit_v2.3.3 with default parameters (Patel and Jain Citation2012). The plastome was de novo assembled using NOVOPlasty (Dierckxsens et al. Citation2017). The complete cp genome was annotated with GeSeq (Tillich et al. Citation2017).
The complete chloroplast genome of O. cordata was a quadripartite circular and 157,879 bp in length (GenBank accession No.: MK697671), comprising a large single copy (LSC) of 87,672 bp and a small single copy (SSC) of 20,265 bp, separated by two inverted repeat (IR) regions of 24,971 bp. The overall GC content was 36.6%. The chloroplast genome contained 72 protein-coding gene, and 37 tRNA genes and 8 rRNA genes. A total of 89 simple sequence repeats (SSRs) were detected using the online software IMEx (Mudunuri and Nagarajaram Citation2007). The numbers of mono-, di-, tri-, tetra-, and penta- nucleotides SSRs are 37, 31, 9, 10, and 2, respectively.
To determine the phylogenetic location of O. cordata, the published chloroplast genomes, including O. acuminata var songmingensis (MK604184, Guo et al. Citation2019) and Elodea canadensis (NC_018541, Huotari and Korpelainen Citation2012) from Hydrocharitaceae and 14 species from Alismatales were used, with 3 Acorus species from Acorales as outgroups. The total 20 chloroplast genomes were aligned using MAFFT 7.308 (Katoh and Standley Citation2013). The maximum likelihood (ML) tree was reconstructed by RAxML 8.2.11 (Stamatakis et al. Citation2008) with the nucleotide substitution model of GTR + G. Bootstrap values were calculated from 1000 replicate analysis. The phylogenetic tree showed that all three sampled species of Hydrocharitaceae formed a monophyletic clade with 100% support. Ottelia cordata and its congeneric species, O. acuminata var. songmingensis, formed a sister clade to Elodea canadensis also with 100% support (). The complete chloroplast genome of O. cordata will provide informative resource both for conservation of this species and for phylogenetic study of Hydrocharitaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Huotari T, Korpelainen H. 2012. Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene. 508:96–105.
- Guo JL, Li ZM, Huang Y, Yu YH, Zhang YH. 2019. The complete chloroplast genome of Ottelia acuminata var. songmingensis, an endangered aquatic macrophyte in China. Mitochondrial DNA Part B. 4:1624–1625.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Mudunuri SB, Nagarajaram HA. 2007. IMEx: imperfect microsatellite extractor. Bioinformatics. 23:1181–1187.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Wang QF, Guo YH, Haynes RR, Hellquist CB. 2010. Hydrocharitaceae. In: Wu ZY, Peter RH, Hong DY, editors. Flora of China. Vol. 23. St Louis: Science Press, Beijing and Missouri Botanical Garden Press; pp. 91–102.
- Yu D, Chong YX, Tu MH, Wang XY, Zhou XH. 1998. Study on the threatened aquatic higher plant species of China. Biodiv Sci. 6:13–21.